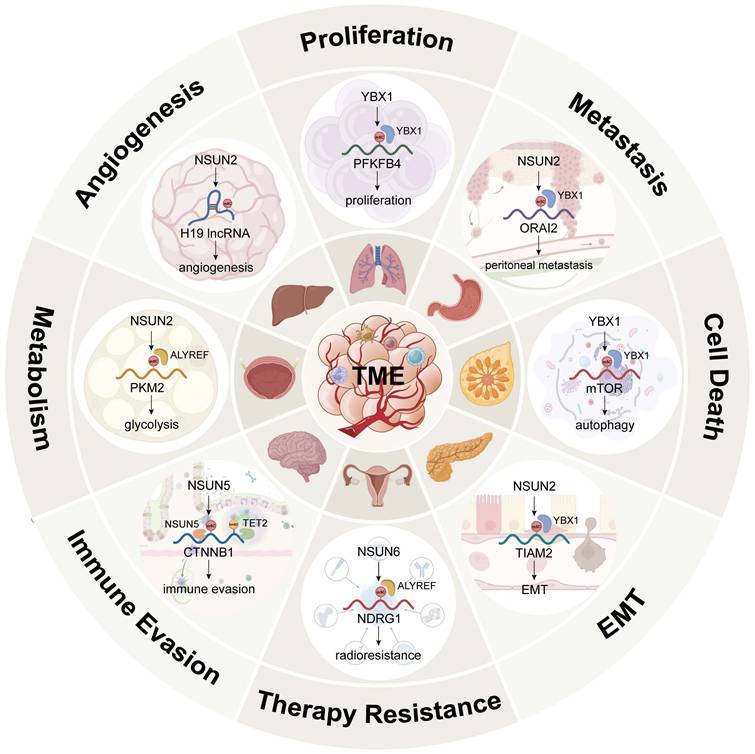
13.3
Impact Factor
Theranostics 2025; 15(16):8404-8428. doi:10.7150/thno.112332 This issue Cite
Review
Epitranscriptomic mechanisms and implications of RNA m5C modification in cancer
1. Department of Gynecology, Xiangya Hospital, Central South University, Changsha, China.
2. Gynecological Oncology Research and Engineering Center of Hunan Province, Changsha, China.
3. National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China.
4. Institute of Medical Sciences, Xiangya Hospital, Central South University, Changsha, China.
*Contributed equally.
Received 2025-2-17; Accepted 2025-7-9; Published 2025-7-25
Abstract
Cancer is an extremely complex disease characterized by abnormal cell growth due to genetic and environmental factors. With the rise of the field of epigenetic transcriptomics, 5-methylcytidine (m5C) modification has been identified as one of the most common chemical modifications occurring in various RNA types. The writers, erasers, and readers of m5C modification regulate cancer initiation, progression, and therapeutic responses, such as the proliferation, metastasis, angiogenesis, metabolic reprogramming, immune escape, and therapeutic resistance of tumour cells, by regulating RNA stability, translation, nuclear export, and splicing processes. In this review, we elucidate the biological process of m5C modification, summarize the abnormal expression of RNA-modifying proteins (RMPs) in common malignant tumours, explore their functional effects on malignant hallmarks of cancer and molecular mechanisms, and prospect the potential clinical application value of m5C.
Keywords: 5-methylcytidine modification, RNA-modifying proteins, cancer, clinical application
1. Introduction
Cancer has become a major public health challenge that threatens human health worldwide. More than 52,900 people are diagnosed with cancer every day, and more than 27,000 people die from it, which places a serious economic burden on society [1]. The initiation of cancer was originally thought to be an entirely genetic disease driven by genes. However, the complexity of cancer reveals that it is a highly structured ecosystem that controls tumour initiation, progression, and therapeutic response [2].
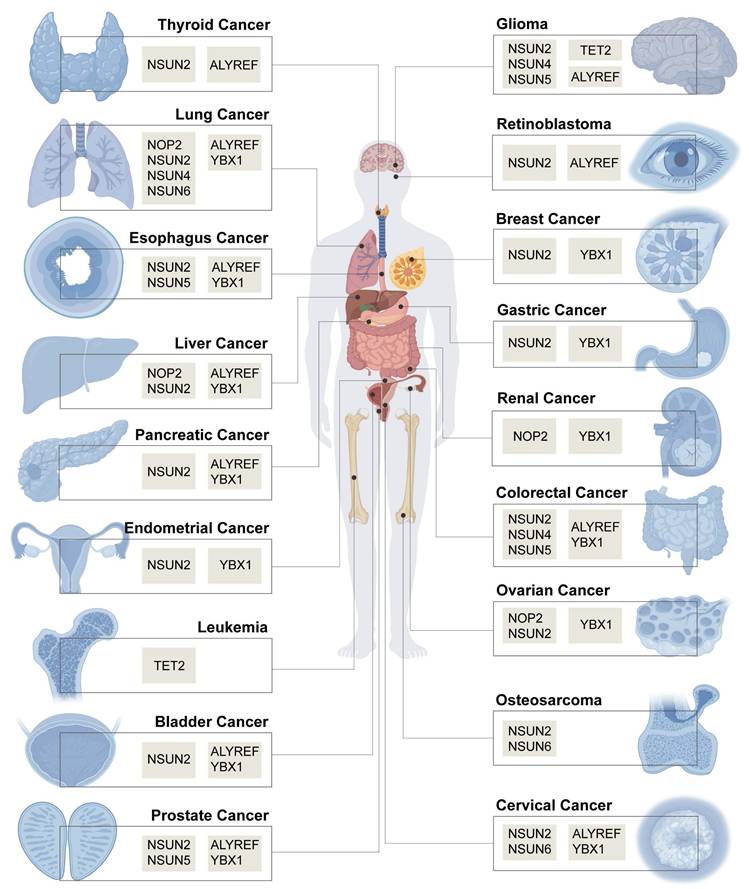
Recently, the discovery of reversible mRNA methylation has opened a new scope of posttranscriptional gene regulation in eukaryotes, especially its role in cancer initiation, progression, and treatment [3-5]. More than 170 RNA modifications have been identified in eukaryotes [6]. Among these modifications, 5-methylcytidine (m5C) RNA modification has attracted increasing attention in cancer research [7]. Initially, m5C modification sites were found mainly in tRNA and rRNA. In 2012, bisulphite sequencing (BisSeq) of whole transcripts in HeLa cells revealed that m5C modification was widely distributed in mRNA and noncoding RNA (ncRNA), and the first transcriptome-wide mapping of m5C in human cells was performed [8]. The roles of TET and ALYREF in m5C were subsequently identified in 2014 and 2017, respectively [9, 10]. An increasing number of studies have shown that m5C is involved in various diseases, such as cardiovascular, liver, Alzheimer, SARS-CoV-2-associated, and autoimmune disease, as well as several cancers [11-14]. Previous excellent reviews have highlighted progress in understanding the role of RNA m5C modification in multiple diseases [13, 15-19]. In addition to discussing newly discovered RNA-modifying proteins (RMPs), m5C modification target RNAs, and updates in our understanding of RNA m5C modification mechanisms and functions in cancers, we summarize the comprehensive functions and molecular mechanisms of RNA m5C modification in more than twenty malignant tumours according to cancer initiation, progression, and therapeutic response. Importantly, we fill gaps in the study of several RMPs in specific cancers, which may provide new ideas for discovering potential biomarkers and therapeutic targets. We also propose the clinical application potential of m5C modification, which lays the foundation for further research.
2. m5C RNA Modification
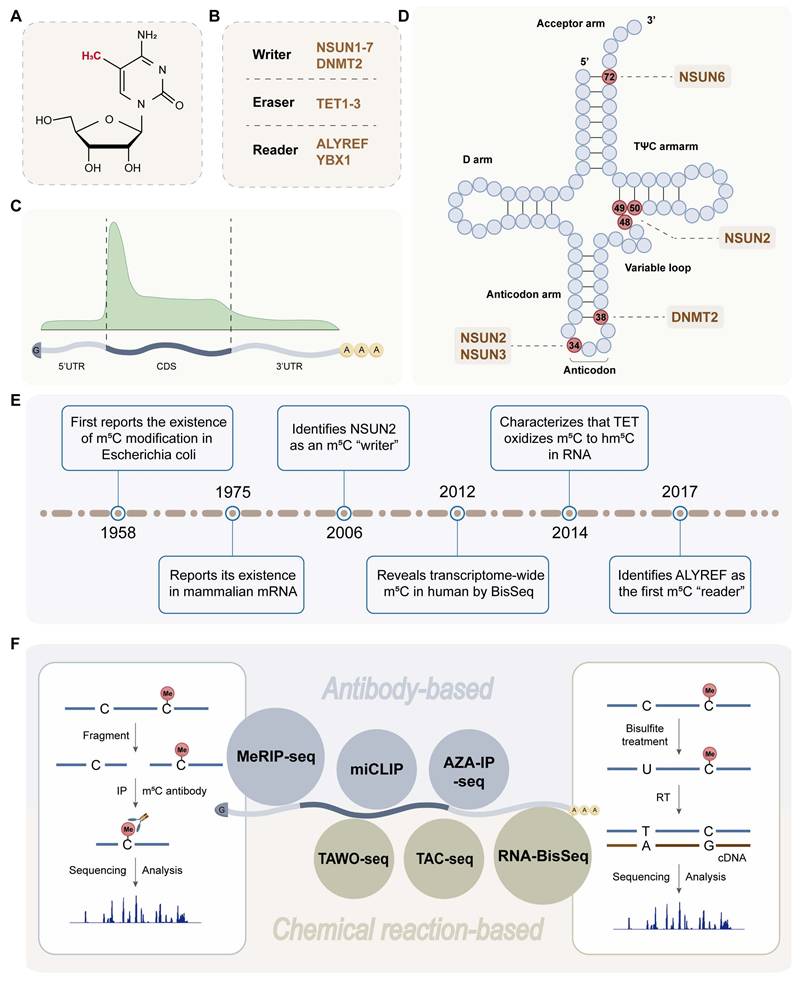
m5C is a chemical modification formed by the addition of a methyl group from the donor, usually S-adenosyl-methionine (SAM), to the fifth carbon atom of cytosine in the RNA molecule [20] (Figure 1A-B). In mammals, m5C modification accounts for approximately 0.02-0.09% of all cytosine modifications [21]. The first cytosine-methylated transcriptome analysis of human cells revealed more than 10,000 m5C sites (>20% methylation) located on approximately 8,500 mRNAs [22], mainly distributed in the coding sequence (CDS) region [23] (Figure 1C). Since m5C was first reported in 1958, this modification has been found to occur in any RNA type, including mRNA, tRNA (Figure 1D), rRNA, and eRNA, among others [17]. Owing to the limitations of m5C modification in coding RNA research, Squires et al. innovatively combined sulphite cell RNA transformation with next-generation sequencing in 2012 [8] (Figure 1E), which promoted our understanding and further exploration of m5C modification in mRNA. Recently, various methods of m5C detection have emerged [24, 25]. There are two main categories, namely, antibody-based and chemical reaction-based methods (Figure 1F). Methylated RNA immunoprecipitation sequencing (MeRIP-seq) uses antibodies specific for m5C or m5C methyltransferase to enrich m5C-modified RNA [26]. Currently, RNA-BisSeq is the most widely used strategy for transcriptome-wide, base-resolution m5C detection [27, 28].
m5C modification is dynamically regulated by three types of RMPs, namely, writers, erasers, and readers [29] (Figure 2). Among them, the main writers include NOL1/NOP2/SUN (NSUN) domain protein family members [30-37] and DNA methyltransferase (DNMT) homologous DNMT2 [38, 39], and the erasers include the TET family (TET1-3) [9, 40, 41] and alkB homologue 1 (ALKBH1) [42, 43]. The m5C sites in RNA are recognized by two main readers, Aly/REF export factor (ALYREF) [10, 44, 45] and Y-box binding protein 1 (YBX1) [46-48], which determine the regulatory mechanism and function of m5C modification in tumour cells. To date, the dysregulated RMPs that have been studied in the field of oncology are shown in Figure 3.
2.1 Writers
m5C RNA methyltransferases (RNMTs) first form a covalent thioester bond, connecting the cysteine residue of their catalytic domain to the C6 position of the target cytosine, forming an RNMT-RNA adduct [49]. Then, RNMTs catalyse the transfer of a methyl group from SAM to the fifth carbon of the cytosine base, forming m5C [50]. NSUN1-7 and DNMT2, as RNMTs, catalyse the m5C modification on different RNAs in different subcellular locations, thereby exerting their respective biological functions.
The human NSUN2 gene is located at 5p15, and its protein has been shown to be localized to the nucleoli situated between or in close proximity to dense heterochromatic regions [51]. The role of NSUN2 is quite extensive, as it acts on a variety of RNA types, such as tRNA, mRNA, and ncRNA. NSUN2-mediated m5C modification of tRNA is common and highly conserved, occurring in the vast majority (>80%) of transcribed tRNA in vivo in humans and mice [52]. Moreover, recognition and methylation by NSUN2 are both site- and structure specific. tRNA contains five conserved domains, including the acceptor arm, the D arm, the anticodon arm, the variable loop (VL) and the TΨC arm (Figure 1D). For eukaryotic tRNA, m5C residues cluster at the junction between the VL and TΨC arms, and C48 and C49 are most frequently modified, with a high prevalence [52]. BisSeq and miCLIP have confirmed robust m5C modification in the anticodon loop at C34 (tRNALeu) and C38 (tRNAAsp, tRNAGly, and tRNAVal) and in the VL junction at C50 (tRNAGlu and tRNAGly) [52, 53]. Notably, methylation at C34/48/49/50 is solely dependent on NSUN2, whereas C38 methylation is mediated by DNMT2 [32, 39, 54]. Mechanistically, NSUN2 protein accommodates the SAM cofactor with its Rossmann-fold catalytic core (residues 171-429) and PUA domain (residues 54-147). In addition, it uses two catalytic cysteines in the active site, which are present in conserved motifs IV (Cys271) and VI (Cys321) [55]. The deposition of m5C at the VL protects tRNA from tRNA‒protein interactions and unnecessary cleavage of mature and functional tRNA during the stress response [55]. In contrast, the functional loss of NSUN2 could result in the absence of tRNA LeuCAA and lead to changes in codon usage, significantly impacting the translation rate of tissue-specific proteins in mammals [56]. NSUN2 is the most important RNA methyltransferase to induce m5C to specific RNAs that regulate the malignant behaviour of various cancers [57-60]. As a cofactor, glucose binds to NSUN2 at amino acids 1-28 to promote NSUN2 oligomerization and activation. Activated NSUN2 increases m5C methylation on TREX2 mRNA and stabilizes TREX2 to restrict cytosolic dsDNA accumulation and cGAS/STING activation to promote tumorigenesis and resistance to anti-PD-L1 immunotherapy [61].
The molecular structure (A), RNA-modifying proteins (B), sites distribution on mRNA (C) and tRNA (D), development history(E) and detection techniques (F) of m5C modification. Created in BioRender. Mao, Z. (2025) https://BioRender.com/6nefran.
The biological processes and molecular mechanisms of m5C function in tumour cells. (A)NSUN2/YBX1/m5C regulates SKIL stability in colorectal cancer. (B)NSUN2/YBX1/m5C regulates QSOX1 translation in lung cancer. (C)NSUN5/ALYREF/m5C regulates ACC1 nuclear export in prostate cancer. (D)NSUN2/ALYREF/m5C regulates RABL6 in bladder cancer. Created in BioRender. Mao, Z. (2025) https://BioRender.com/n3el9cc.
NSUN1 (NOP2) is characterized primarily in budding yeast as an essential ribosomal biogenesis factor required for the deposition of m5C on 25S rRNA [31]. miCLIP-seq has revealed that rRNA is the major m5C-specific target of NSUN1 in human cells. Human NSUN1 binds to the rRNA 5′-ETS region and crosslinks to 28S rRNA at position C4447 [62]. NSUN3 initiates m5C biogenesis at position C34 in human mitochondrial tRNAMet, regulating mitochondrial protein synthesis, oxygen consumption, and mitochondrial activity [33]. Mitochondrial m5C modification is essential for the dynamic regulation of mitochondrial translation rates and thereby shapes metabolic reprogramming during tumour metastasis [63]. NSUN4 methylates cytosine 911 in the 12S rRNA of the small subunit (SSU), playing a key role in controlling the final step of ribosomal biogenesis to ensure that only the mature SSU and large subunit (LSU) are assembled [34, 64]. Similarly, NSUN5 acts as an RNA methyltransferase at the C3782 position of human 28S rRNA, which regulates the adaptive translational program for survival under conditions of cellular stress [65]. Human NSUN6 is associated with tRNA and acts as a tRNA methyltransferase. It can catalyse cytosine 72 at the 3' end of tRNACys and tRNAThr [66].
Although the sequence and structure of DNMT2 (also known as TRNMT1) have close affinities for authentic DNA cytosine methyltransferases, the substrate of the highly conserved human DNMT2 was found to be predominantly aspartic acid-transfer RNA because the presence of a DNA competitor weakens but cannot eliminate the DNMT2-RNA complex signal [39, 67]. An increasing number of studies have shown that DNMT2 and its homologues can modify C38 of tRNAGly, tRNAAsp, tRNAGlu, and tRNAVal in vivo in mammals and other species [68-71]. In contrast with the NSNU family, only DNMT2 methylates RNA by utilizing a single cysteine (Cys79) in its catalytic pocket and through a DNMT-like catalytic mechanism [72]. tRNA methylation catalysis by human DNMT requires C79 in motif IV (PCQ), E119 in motif VI (ENV), and R160 and R162 in motif VIII (RXR). In addition, some residues (such as I228, Q221, L229, G305, and Y297) located on the surface of the target recognition domain (TRD) and target recognition extension domain (TRED) regions in DNMT2 contribute to the selection of preferred substrate tRNA [72, 73]. Importantly, DNMT and NSUN2 exhibit complementary target site specificities and collaborate to facilitate tRNA methylation by complementing each other in terms of gene expression, promoting tRNA stability and accurate protein synthesis [68, 74-76]. DNMT2 is widely involved in a variety of physiological regulatory processes. For example, DNMT2 deletion increases cancer cell sensitivity to radiation and PARP inhibitors (PARPis). This role is dependent on its m5C writer effect [77]. DNMT2-dependent m5C in damage-induced R-loops promotes transcription coupled-homologous recombination (TC-HR) and simultaneously suppresses PARP1-mediated alternative nonhomologous end joining (Alt-NHEJ), ensuring that TC-HR is the preferred double-strand break (DSB) repair pathway in transcribed regions [78]. Overall, previous studies have shown that the above RNMTs function primarily on tRNA and rRNA. Interestingly, they have been shown to be associated with mRNA methylation [79].
The m5C RNA-modifying proteins that play a key role in cancer. Created in BioRender. Mao, Z. (2025) https://BioRender.com/eu8anj3.
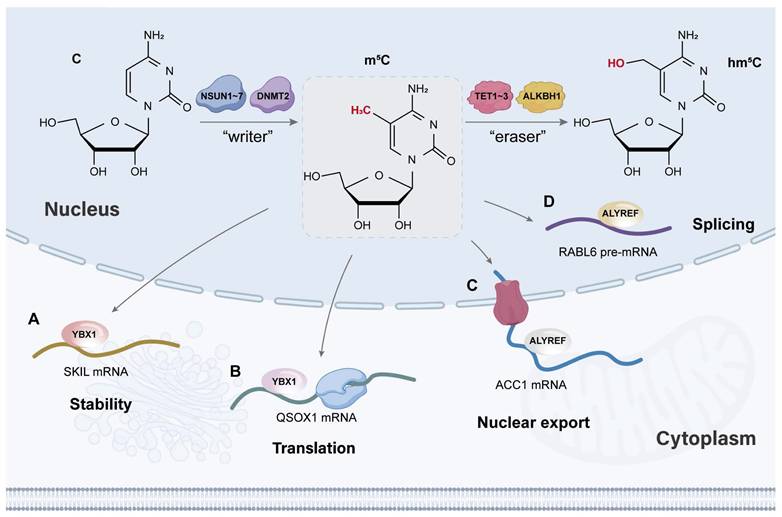
2.2 Erasers
As a dynamic process, added methyl groups can be removed by demethylases (erasers). Previously, the reversible biological process of m5C modification has remained controversial. Over the years, m5C has been shown to be oxidized by TET1-3 and ALKBH1 to bioactive 5-hydroxymethylcytosine (hm5c) [9, 42, 63, 80].
TET family members were initially identified as DNA demethylases for a variety of nucleic acid substrates [81]. The primary structure of TET enzymes includes a carboxy-terminal catalytic domain composed of a cysteine-rich domain (CRD) and two double-stranded β-helix (DSBH) regions flanking an extended low-complexity insertion region [82]. The DSBH domains harbour conserved residues critical for coordinating cofactors (Fe(II) and α-ketoglutarate) essential for catalysis. The catalytic core is stabilized by two zinc finger motifs that structurally integrate the DSBH regions with the CRD, forming a compact functional module. This architecture ensures proper spatial alignment of cofactor-binding sites and catalytic residues, enabling the oxidative modification of methylated cytosines during demethylation [83, 84]. Interestingly, Fu et al. reported that TETs could also participate in the dynamic and reversible modification of RNA cytosines [9]. Multiple studies have indicated that TET1 and TET2 are required for the deposition of 5hmC in mRNA and tRNA and that TET-mediated 5hmC can reduce the stability of important pluripotency-promoting transcripts during embryonic stem cell (ESC) differentiation [9, 85-88]. Importantly, a proteomic approach confirmed that TET1 and TET2 contain an RNA-binding domain [88]. TET1-mediated m5C RNA modification, demethylation, and R-loop resolution during DNA repair are important for repair completion and the maintenance of genome stability [89]. TET2 mutations with high frequency have been identified in multiple haematologic malignancies [90-93]. The relationship between TET2 mutations and overall survival suggests that TET2 functions as a tumour suppressor [94, 95]. For example, TET2 regulates the open state of active chromatin by oxidizing the m5C modification of caRNA and inhibits leukaemogenesis [96]. Surprisingly, several findings on therapeutic resistance also support the tumour-promoting role of TET2 [97-99].
ALKBH1, which is widely distributed in the cytoplasm, nucleus, and mitochondria, has substrate diversity and can remove multiple types of RNA modifications, such as N1-methyladenosine (m1A), m6A, m5C, and 3-methylcytidine (m3C) [100-102]. It contains a central catalytic core, a nucleotide recognition lid (NRL) with Flip1 and Flip2, and a distinct N-terminal Flip0. In the catalytic core, there is a highly conserved DSBH structure [102]. Notably, three unique structural features outside the core determine the high dependence of ALKBH1 on the secondary structure of the substrate. Specifically, ALKBH1 preferentially catalyses demethylation in bulged, bubbled DNA and various local unpaired nucleic acids (such as R-loops, stem loops, D-loops, and bulges) [103]. Compared with TET2, ALKBH1 is the major m5C dioxygenase of RNA in human HEK293T cells, where it is responsible for the bulk of hm5C and f5C production [42]. Hypoxia-induced ALKBH1 decreases the global m5C level in human extravasated trophoblast cells and can regulate mRNA stability [104]. In addition, human ALKBH1 catalyses the hydroxylation and oxidization of m5C34 in both ct-tRNALeu and mt-tRNAMet, affecting mitochondrial translation and respiratory complex activity [104]. To date, ALKBH1 has not been found to function as an RNA demethylase in malignant tumours.
2.3 Readers
Reader proteins, with special RNA-binding domains, are the ultimate executors of RNA methylation functions (Figure 2).
ALYREF, which is located mainly in the cell nucleus, is the first mRNA m5C-reading protein with the critical m5C recognition site K171 to be discovered, and it preferentially binds mature mRNA globally [10]. As a component of the TREX complex, it facilitates the nuclear export of mRNA by specifically binding to mRNA with m5C modifications in the nucleus to form the mRNA-exporting protein (mRNP) complex [45, 105, 106]. Mechanistically, CBP80 and PABPN1 are specifically involved in ALYREF recruitment to the 5′ and 3′ regions of mRNA. Moreover, CstF64 interacts with ALYREF and functions in ALYREF recruitment to the mRNA [105]. Studies have suggested that ALYREF can play an essential role in metastasis, cancer progression, and chemoresistance by modulating cell proliferation, migration, and invasion and antiapoptotic effects [45, 107-110].
YBX1 is localized primarily in the cytoplasm and serves as an RNA m5C reader that plays a crucial role in regulating RNA metabolism [111]. YBX1 comprises three primary structural domains: the cold shock domain (CSD), the alanine/proline domain (A/P domain), and the C-terminal domain (CTD) [112]. These domains mediate complex interactions between YBX1 and both nucleic acids (DNA and RNA) and other proteins. The CSD uniquely contains nucleic acid-binding sites, enabling YBX1 to engage preferentially with m5C-modified RNA. High-resolution crystal structures have revealed that CSD interacts with RNA mainly through π-π stacking interactions assembled by four highly conserved aromatic residues (His-87, Phe-85, Phe-74, and Trp-65) [48, 113]. Subsequently, it promotes mRNA stability in an m5C-dependent manner by recruiting the mRNA stabilizer ELAVL1 [47]. YBX1 is expressed in a broad range of tissues, and its roles in regulating cell proliferation, stress responses, and apoptosis make it crucial for normal development and tissue homeostasis [114, 115]. In addition, its dysregulation has been linked to various diseases, including cancer [116-119].
3. The Fates of RNA Molecules with m5C Modifications
3.1 mRNA
Generally, m5C modification participates in four metabolic processes of mRNA, including pre-mRNA splicing, nuclear export, stability, and translation of mature mRNA, thereby altering the expression of m5C-related genes [10, 118, 120-122] (Figure 2). Taking the most common RNMP as an example, NSUN2 induces different biological mechanisms in various mRNAs. On the one hand, NSUN2 alters the methylation pattern of PTEN pre-mRNA, resulting in the downregulation of PTEN expression by mediating its alternative splicing events [123]. On the other hand, NSUN2 functions as a writer of m5C modifications on SRSF6 mRNA. The increasing level of m5C induce its nuclear‒cytoplasmic transport, which plays a vital role in multidrug resistance. In addition, NSUN2-mediated m5C modification enhances FABP5 and LAMC2 stability in osteosarcoma (OS) and head and neck squamous cell carcinoma (HNSCC), respectively [124, 125]. Moreover, the overexpression of wild-type NSUN2 leads to gefitinib resistance and tumour recurrence, which are related to the m5C site at the CDS region of QSOX1 mRNA. Interestingly, unlike others, the increasing m5C modification of QSOX1 promotes its translation [126].
3.2 tRNA
tRNA shows the widest variety and largest number of RNA modifications, which are pivotal for stabilizing the tertiary structure of tRNA molecules (modifications outside of the anticodon loop) and decoding the genetic code (modifications in the anticodon loop). NSUN2 is upregulated in anaplastic thyroid cancer (ATC) and increases the m5C modification on tRNAleu at the C48 site, which stabilizes tRNAleu by preventing its cleavage. This stable tRNAleu maintains homeostasis and rapidly transports leucine, substantially increasing the efficiency necessary to support the translation of c-MYC, BCL2, RAB31, JUNB, and TRAF2, among others. As pro-oncogenic proteins, they contribute to promoting tumour formation, proliferation, invasion, migration, and resistance to genotoxic drugs [127].
3.3 circRNA
circRNA is a class of covalently closed RNA molecules characterized by universality, diversity, stability and conservative evolution. Recent studies have shown that some epitranscriptomic modifications affect circRNA metabolism, such as stability, subcellular localization, and even translation. The m5C modification of circFAM190B increases its stability, which is dependent on NSUN2. circFAM190B targets SFN and regulates its ubiquitination, thereby inhibiting cellular autophagy through the SFN/mTOR/ULK1 pathway and ultimately promoting lung cancer development [128]. The increased circ_0102913 expression in cancer cells was attributed to NSUN5 at least partly because the hypermethylated m5C modification stabilizes the specific RNA. It subsequently enhances the malignant properties of cells via the miR-571/RAC2 axis [129]. The carcinogenic effects of RAC2 might be attributed to its role in the alternative activation of macrophages [130]. A combined m5C microarray analysis revealed that circERI3 contains m5C modifications and that the NSUN4-mediated m5C modification of circERI3 could increase its nuclear export. Additionally, circERI3 inhibits DDB1 ubiquitination and regulates PGC-1α transcription through DDB1, thus increasing mitochondrial energy metabolism and ultimately contributing to the development of lung cancer [131].
3.4 lncRNA
lncRNA plays two distinct roles in epitranscriptomic modifications. On the one hand, lncRNA has emerged as a critical regulator of RMPs. In addition, there are many sites on their sequences that can be modified. In glioblastoma endothelial cells, NSUN2 increases the stability of LINC00324 and upregulates its expression through m5C modification. LINC00324 competes with the 3′-UTR of CBX3 mRNA for binding to the AUH protein and reducing CBX3 mRNA degradation. In addition, CBX3 directly binds to the promoter region of VEGFR2, enhancing VEGFR2 transcription and promoting angiogenesis [132]. The stable lncRNA NR_033928 with m5C modification can upregulate the expression of glutaminase by interacting with the IGF2BP3/HUR complex, which is a potential prognostic and therapeutic target in gastric cancer [133]. The expression of H19 lncRNA is abnormally increased in liver cancer, and this RNA is a specific target of NSUN2. Through m5C modification, its stability is significantly increased, and it recruits the oncoprotein G3BP1, further leading to the accumulation of MYC, which is a new mechanism of angiogenesis [134].
4. Functions and Mechanisms of m5C Modification in Cancer
To date, a total of 14 cancer hallmarks have been identified to explain the mechanisms of malignant tumour initiation, progression, and therapeutic response [135, 136]. Among them, nonmutational epigenetic reprogramming, defined as enabling characteristics, was officially shown to play a significant role in 2022 [137]. Figure 4 and Table 1 summarize the functions and regulatory mechanisms of m5C in cancer.
The functions and mechanisms of m5C in cancer. Created in BioRender. Mao, Z. (2025) https://BioRender.com/e9rc6z6.
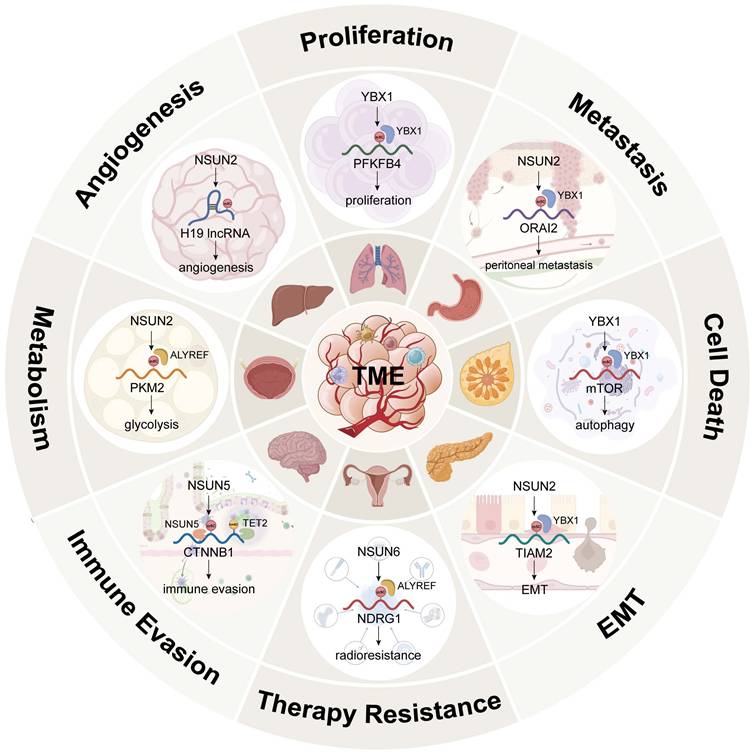
The functions and mechanisms of m5C RMPs in cancer.
| Type | RMPs | Target RNA | Mechanism | Function | Ref. | |||
|---|---|---|---|---|---|---|---|---|
| Glioma | Writer | NSUN2 | Up | ATX | Nuclear export, Translation (ALYREF) | ATX-LPA axis | Migration | [139] |
| LINC00324 | Stability | CBX3-VEGFR2 axis | Proliferation, Migration | [132] | ||||
| NSUN4 | Up | CDC42 | Stability (ALYREF) | PI3K-AKT signaling | Proliferation, Migration, Invasion | [138] | ||
| NSUN5 | Down | CTNNB1 | Stability | TET2-RBFOX2 axis | Immune evasion | [143] | ||
| HNSCC | Writer | NSUN2 | Up | LAMC2 | Stability (YBX1) | - | Proliferation, Migration, Invasion, EMT | [125] |
| Retinoblastoma | Writer | NSUN2 | Up | PFAS | Stability (ALYREF) | - | Proliferation | [4] |
| Nasopharyngeal carcinoma | Reader | ALYREF | Up | NOTCH1 | Stability (ALYREF) | Notch signaling | Proliferation, Migration, Invasion | [153] |
| Thyroid cancer | Writer | NSUN2 | Up | SRSF6 | Nuclear export (ALYREF) | UAP1-AGX2-ABC transporter axis | Multidrug resistance | [108] |
| tRNALeu | Stability | c-MYC/BCL2/RAB31/JUNB/TRAF2 | Proliferation, Migration, Invasion, Chemotherapy resistance | [127] | ||||
| Esophageal cancer | Writer | NSUN2 | Up | GRB2 | Stability | PI3K-AKT and ERK/MAPK signaling | Proliferation, Migration, Invasion | [157] |
| SMOX | Stability (YBX1) | mTORC1 signaling | Proliferation, Migration, Invasion | [156] | ||||
| NLRP3 | Nuclear export, Stability (ALYREF, YBX1) | NLRP3/caspase 1/IL-1β inflammatory pathway | Proliferation, Migration, Invasion | [121] | ||||
| NSUN5 | Up | METTL1 | - | - | Proliferation | [158] | ||
| Reader | ALYREF | Up | TBL1XR1, KMT2E | Stability | Upregulate APOC1 expression | Oxaliplatin resistance | [163] | |
| YBX1 | Up | CSF2 | Stability | - | Migration, Invasion, Glycolysis | [167] | ||
| Breast cancer | Writer | NSUN2 | Up | HGH1 | Stability, Translation (YBX1) | Bind to EEF2 | Proliferation, Migration, Invasion | [252] |
| Reader | YBX1 | Up | mTOR | Stability | - | Proliferation, Migration, Autophagy | [253] | |
| Lung cancer | Writer | NOP2 | Up | EZH2 | Stability (ALYREF) | H3K27me3-E-cadherin axis | Migration, Invasion, EMT | [147] |
| NSUN2 | Up | QSOX1 | Translation (YBX1) | - | EGFR-TKIs resistance | [126] | ||
| NRF2 | Stability (YBX1) | Enhance the transcription of GPX4, FTH1, and other antioxidants | Proliferation, Migration, Invasion, Ferroptosis | [145] | ||||
| PD-L1 | Stability (ALYREF) | Inhibit CD8+ T-cell infiltration | Immune evasion | [146] | ||||
| CircFAM190B | Stability | SFN-mMOR-ULK1 axis | Proliferation, Migration, Apoptosis | [128] | ||||
| ME1, GLUT3, CDK2 | Stability (ALYREF) | - | Proliferation, Migration, Invasion, Angiogenesis, Cell cycle, Metabolism | [109] | ||||
| NSUN4 | Up | CircERI3 | Nuclear export | DDB1-PGC-1α-mitochondria axis | Mitochondrial energy metabolism, Proliferation, Migration, Cell cycle, Apoptosis | [131] | ||
| CDC20 | Stability (ALYREF) | - | Proliferation, Migration, Invasion | [110] | ||||
| NSUN6 | Down | NM23-H1 | - | - | Proliferation, Migration, EMT | [148] | ||
| Reader | ALYREF | Up | YAP1 | Stability | Hippo and Wnt/β-catenin signaling | Proliferation, Migration, Invasion, Apoptosis, Cell cycle, Therapy resistance | [149] | |
| YBX1 | Up | PFKFB4 | Stability | - | Proliferation, Migration, Glycolysis | [118] | ||
| Gastric cancer | Writer | NSUN2 | Up | NR_033928 | Stability | HUR/IGF2BP3-GLS axis | Proliferation, Apoptosis, Glutamine metabolism | [133] |
| NTN1 | Stability | - | Migration, Invasion, Neural invasion | [172] | ||||
| ORAI2 | Stability (YBX1) | PI3K-AKT signaling | Proliferation, Migration, Invasion, Peritoneal metastasis | [173] | ||||
| PIK3R1, PCYT1A | - | - | Proliferation, Migration, Invasion, Chemotherapy resistance | [5] | ||||
| FOXC2 | Stability (YBX1) | - | Proliferation, Migration, Invasion | [174] | ||||
| PTEN | Splicing | PI3K-AKT signaling | Proliferation, Migration | [123] | ||||
| ATG9A | Stability (YBX1) | - | 5-Fluorouracil resistance, Autophagy | [265] | ||||
| Liver cancer | Writer | NOP2 | Up | c-MYC | Stability, Translation | LDHA/PKM2/ENO1/TPI1 | Glycolysis | [180] |
| NSUN2 | Up | GRB2, RNF115, AATF | - | Ras signaling | Sorafenib resistance | [181] | ||
| H19 | Stability | Recruit the G3BP1 oncoprotein | Proliferation, Migration, Invasion, Angiogenesis | [134] | ||||
| SREBP2 | Stability (YBX1) | - | Proliferation, Migration, EMT, Cholesterol metabolism | [119] | ||||
| PKM2 | Stability | - | Proliferation, Migration, Glycolysis | [182] | ||||
| MALAT1 | Stability (ALYREF) | ELAVL1-SLC7A11 axis | Ferroptosis, Sorafenib resistance | [267] | ||||
| SOAT2 | Stability | - | Proliferation, Migration, Invasion, Apoptosis, Immune evasion | [58] | ||||
| Reader | ALYREF | Up | EGFR | Stability | STAT3 signaling | Proliferation, Migration, Invasion, EMT | [177] | |
| YBX1 | Up | RNF115 | Circularization, Translation | DHODH K27 ubiquitination | Ferroptosis | [178] | ||
| Cholangiocarcinoma | Writer | NSUN5 | Up | GLS | Stability | - | Proliferation, Migration, Invasion, Cuproptosis | [186] |
| Reader | ALYREF | Up | PKM2 | Stability | - | Proliferation, Migration, Glycolysis, Ferroptosis | [120] | |
| Pancreatic cancer | Writer | NSUN2 | Up | TIAM2 | Stability (YBX1) | - | Proliferation, Migration, Invasion, EMT | [196] |
| Reader | YBX1 | Up | EGR1, NTRK1, SMAD7 | Stability | MIF/TNF-α | Perineural invasion | [192] | |
| Caspase-8 | Stability | PIPK1/PIPK3/MLKL pathway | Proliferation | [193] | ||||
| ALYREF | Up | JunD | Stability | SLC7A5-mTORC1 signaling | Proliferation, Immune evasion | [194] | ||
| Colorectal cancer | Writer | NSUN2 | Up | ENO1 | Stability (YBX1) | - | Proliferation, Invasion, Glycolysis | [197] |
| SKIL | Stability (YBX1) | Activate TAZ expression | Proliferation, Migration | [198] | ||||
| SLC7A11 | Translation, Stability | - | Proliferation, Ferroptosis | [199] | ||||
| KSR1 | Stability (YBX1) | ERK/MAPK signaling | Migration, Invasion | [59] | ||||
| NSUN4 | Up | NXPH4 | Stability | PHD4-HIF1A axis | Proliferation, Migration, Invasion, RNautophagy | [200] | ||
| NSUN5 | Up | circ0102913 | Stability | miR-571-RAC2 axis | Proliferation, Migration, Invasion | [129] | ||
| GPX4 | Stability | cGAS-STING signaling | Anticancer immunity | [208] | ||||
| Reader | ALYREF | Up | RPS6KB2, RPTOR | Nuclear export | - | Proliferation, Migration | [211] | |
| Renal cancer | Writer | NOP2 | Up | APOL1 | Stability (YBX1) | PI3K-AKT signaling | Proliferation, Migration, Invasion | [213] |
| Reader | YBX1 | Up | PEBP1 | Stability | - | Migration, Invasion | [215] | |
| Bladder cancer | Writer | NSUN2 | Up | RABL6, TK1 | Splicing, Stability (ALYREF) | - | Proliferation, Invasion | [218] |
| HDGF | Stability (YBX1) | - | Proliferation, Migration, Invasion | [47] | ||||
| Reader | ALYREF | Up | PKM2 | Stability | - | Glycolysis | [220] | |
| Prostate cancer | Writer | NSUN2 | Up | AR | Stability (YBX1) | - | Proliferation, Migration, Invasion | [226] |
| TRIM28 | Stability | - | Proliferation, Migration | [60] | ||||
| NSUN5 | Up | ACC1 | Nuclear export (ALYREF) | - | Proliferation, Lipid deposition | [223] | ||
| Ovarian cancer | Writer | NOP2 | Up | RAPGEF4 | - | - | Proliferation, Migration, Invasion | [247] |
| NSUN2 | Up | E2F1 | Stability (YBX1) | MYBL2/RAD54L | Proliferation, Migration, Invasion | [246] | ||
| Reader | YBX1 | Up | CDH3 | Stability | HR-related proteins, such as BRCA1, RAD50, NBS1, RAD51, etc. | Apoptosis, Cisplatin resistance | [242] | |
| E2F5, YY1, RCC2 | Stability | - | Proliferation, Migration, Invasion, Chemoresistance | [243] | ||||
| Endometrial cancer | Writer | NSUN2 | Up | SLC7A11 | Stability (YBX1) | - | Ferroptosis resistance | [239] |
| Cervical cancer | Writer | NSUN2 | Up | LRRC8A | Stability (YBX1) | PI3K-AKT signaling | Proliferation, Migration, Invasion | [233] |
| KRT13 | Stability (YBX1) | Migration, Invasion | [234] | |||||
| NSUN6 | Up | NDRG1 | Stability (ALYREF) | HR-mediated DNA damage repair | Radioresistance | [232] | ||
| Leukemia | Writer | NSUN2 | Up | PHHGH, SHMT2 | Stability (YBX1) | - | Proliferation, Apoptosis, Serine metabolism | [263] |
| Eraser | TET2 | Down | TSPAN13 | Stability (YBX1) | - | stem cell homing and self-renewal | [93] | |
| Melanoma | Writer | NSUN2 | Up | CTNNB1 | - | c-MYC/Cyclin D1 | Proliferation, Migration | [144] |
| Reader | YBX1 | Up | MAGEA1 | Stability | P53 signaling | Stemness, Proliferation, Migration, Invasion | [268] | |
| Osteosarcoma | Writer | NSUN2 | Up | FABP5 | Stability | - | Proliferation, Migration, Invasion, Fatty acid metabolism | [124] |
| NSUN6 | Up | EEF1A2 | Stability | AKT/mTOR signaling | Proliferation, Migration, Invasion | [122] | ||
4.1 Central nervous system cancers
4.1.1 Glioma
The level of m5C modification in glioma tissue is significantly greater than that in peritumoral tissue and is positively correlated with the tumour grade [138]. While NSUN2 and NSUN4 are highly expressed in glioma, single-cell bioinformatic analysis has revealed that malignant cells present the lowest NSUN5 expression levels among the different cell types that make up the tumour mass. NSUN2 methylates the 3′-UTR of ATX mRNA at the C2756 site in the human glioma cell line U87. With the recognition of ALYREF, ATX is exported from the nucleus to the cytoplasm and subsequently translated into ATX protein [139]. ATX is a secreted glycoprotein that can convert lysophosphatidylcholine into lysophosphatidic acid (LPA), functioning as the major enzyme for extracellular LPA production. LPA can regulate a broad range of cell functions, such as cell survival, proliferation, and migration [140, 141]. Malignant gliomas exhibit immune evasion characterized by increased expression of the immune checkpoint protein CD47 [142]. By combining databases, the m5C prediction website, and the MeRIP-qPCR assay, Zhao et al. revealed for the first time that NSUN4, as a key writer for controlling m5C levels in glioma, mediates changes in m5C levels to promote the stability of CDC42 mRNA. This cascade, in turn, promotes activation of the PI3K-AKT pathway, culminating in the malignant progression of glioma cells [138]. NSUN5 directly interacts with CTNNB1 caRNA and increases its m5C modification, which is subsequently oxidized by TET2 to 5hmC. RBFOX2 functions as a 5hmC-specific reader to recognize and promote CTNNB1 degradation. Finally, the downregulation of β-catenin interferes with the binding of CD47 to SIRPα, thereby weakening the phagocytosis of tumour-associated macrophages (TAMs) [143]. Intriguingly, this study revealed that NSUN5 could act as an immune therapy target to transform glioma into a “warm tumour” and lead to impressive therapeutic outcomes when NSUN5 is restored in IDH1-R132H mutant glioma cells.
3.1.2 Ocular cancer
The global and mRNA m5C levels are significantly enriched in retinoblastoma (RB) tissue compared with normal retinal tissue, which is attributed to the high expression of tumour-specific NSUN2 [4]. Through multiomic analysis, PFAS mRNA has been identified as a downstream candidate target of NSUN2. As a vital enzyme in purine biosynthesis, PFAS upregulated by m5C modification accelerates retinoblastoma progression, which bridges the current understanding of RNA modification and metabolic reprogramming. NSUN2 increases m5C modification on CTNNB1 mRNA and then promotes uveal melanoma cell migration and proliferation by regulating the cell cycle [144].
4.2 Respiratory tract cancers
4.2.1 Lung cancer
Lung cancer remains the leading cause of cancer-related deaths worldwide, and the most prevalent histological type is non-small cell lung cancer (NSCLC), which constitutes approximately 80% of all cases. NOP2, NSUN2, and NSUN4, key RNA m5C methyltransferases, are highly expressed in NSCLC tumour tissue, and their levels are strongly correlated with tumour grade, tumour size, and poor outcomes. In addition, ALYREF and YBX1, which are readers of m5C, are upregulated in lung cancer. However, the levels of NSUN6 are low in lung cancer, and NSUN6 may play a protective role. Cr(VI), a common environmental contaminant, has been shown to result in NSUN2 upregulation in human bronchial epithelial cells and mouse lung tissues [109]. Using RNA-seq, MeRIP-seq, and MeRIP-qPCR, several targets of NSUN2 have been identified, including mRNAs (QSOX1, NRF2, PD-L1, ME1, GLUT3, and CDK) [109, 126, 145, 146] and circRNAs (circFAM190B) [128]. Chen et al. reported that NSUN2-mediated m5C modification of the NRF2 mRNA 5′-UTR enhances its stability in an m5C-YBX1-dependent manner. NRF2 is renowned for its integral role in managing ferroptosis, which relies on the disengagement of KEAP1 from NRF2 when faced with oxidative stress [145]. Interestingly, the findings of m5C modification have shed light on a novel, noncanonical pathway in which NRF2 activation modulated by NSUN2 operates independently of the KEAP1-mediated mechanism. In contrast, NSUN2 posttranscriptionally enhances PD-L1 mRNA stability, subsequently increasing PD-L1 expression in an m5C-ALYREF-dependent manner and providing protective effects for tumour cells against CD8+ T-cell-mediated cytotoxicity in NSCLC [146]. Additionally, NOP2 and NSUN4 are highly expressed in lung cancer. The stable EZH2 mRNA produced by NOP2 and ALYREF coregulation leads to EMT and promotes the malignant properties of cancer cells through the H3K27me3/E-cadherin axis [147]. NSUN6 regulates NM23-H1 expression by modifying the 3′-UTR of NM23-H1 mRNA through the m5C mechanism and inhibits cancer cell proliferation, migration, and EMT [148]. These studies have greatly enriched the understanding of the role of writers other than NSUN2 in cancer regulation. The binding of ALYREF to YAP1 mRNA inhibits the apoptosis of tumour cells through activation of the Hippo and Wnt/β-catenin pathways [149]. YBX1 ensures the stability of PFKFB4 mRNA by recognizing its 3′-UTR m5C sites in the cytoplasm after the exportation effect of THOC3 [118]. PFKFB4, a glycolysis regulator, produces pentose phosphate to perform carcinogenic functions [150].
4.2.2 Nasopharyngeal carcinoma
Nasopharyngeal carcinoma (NPC) has high rates of metastasis and invasion, with a particularly high incidence in Southeast Asia, southern China, and North Africa [151, 152]. NSUN2 and ALYREF are significantly upregulated in NPC tissues, and their high expression is correlated with poor distant metastasis-free survival (DMFS) and overall survival (OS) [107, 153]. The analysis of GSEA RNA-seq data revealed that NOTCH1 mRNA is m5C-modified by NSUN2. This protein is subsequently recognized and stabilized by ALYREF, which promotes NOTCH1 expression and activates the Notch signalling pathway in NPC cells. Notably, the evolutionarily conserved Notch signalling pathway plays an important role in determining the fate of NPC cells. Moreover, treatment with the NOTCH1 inhibitor LY3039478 and its relationship with prognosis in this study highlighted that ALYREF could serve as a therapeutic target and potential biomarker.
4.3 Digestive tract cancers
4.3.1 Esophageal cancer
Esophageal squamous cell carcinoma (ESCC) is one of the most aggressive gastrointestinal malignancies worldwide, with a 5-year survival rate of approximately 20% [154, 155]. The levels of RNA m5C methylation are substantially increased in ESCC tissues due to the upregulation of NSUN2 and NSUN5, which constitutes an important regulatory mechanism for ESCC progression [156-158]. Additionally, ALYREF and YBX1 levels are also elevated in ESCC. NSUN2 increases the m5C modification on GRB2 and SMOX mRNA and promotes their stability [156, 157]. The upregulation of GRB2 evokes oncogenic PI3K/AKT and ERK/MAPK signalling [159]. SMOX activates the mTORC1 signalling pathway with the recognition of YBX1. NSUN5 is also significantly upregulated in esophageal Cancer (EC) and shows promising diagnostic potential [158]. Gene coexpression analysis of data from the databases GEPIA and UALCAN and site analysis from RMBase v3.0 have suggested that NSUN5 binds directly to the METTL1 transcript, facilitating its m5C modification in EC cells. METTL1, an m7G-modifying enzyme, has been identified as a novel epigenetic oncogene, and elevated METTL1 activity is essential for promoting EC tumour growth [160, 161]. Oxaliplatin (L-OHP) is a potent chemotherapeutic agent that induces apoptosis in EC cells [162]. However, its effectiveness is significantly hindered by the development of resistance. ALYREF expression is elevated in L-OHP-resistant EC tissues, and ALYREF further recognizes the m5C sites on TBL1XR1 and KMT2E mRNAs, stabilizing these transcripts and promoting APOC1 expression [163]. APOC1, a protransfer factor, plays a crucial role in the metabolism of very-low-density lipoprotein (VLDL) and high-density lipoprotein (HDL) cholesterol, predicting a poor prognosis and correlating with tumour immune infiltration [164-166]. Within the cytoplasmic milieu of ESCC cells, circPRKCA interacts with YBX1, consequently preventing the ubiquitination-mediated degradation of YBX1. Increased concentrations of YBX1 increase the stability of CSF2 mRNA in a m5C-dependent manner [167]. CSF2, a tumour-derived growth factor, is widely recognized for its role in promoting angiogenesis, which is often a crucial process [168]. Additionally, it drives EMT and enhances immune checkpoint protein expression, thereby facilitating the malignant progression of cancer [169, 170]. Hence, these findings highlight the potential of RMPs as more comprehensive biomarkers due to the broader involvement of RMPs in the critical pathways and tumorigenesis of EC, providing a preclinical rationale for selectively targeting m5C modification as a promising therapeutic strategy.
4.3.2 Gastric cancer
Gastric cancer (GC) is the fifth most common malignant tumour and the fourth leading cause of cancer-associated death worldwide [171]. Overall, the RNA m5C content is increased in GC samples and is positively correlated with NSUN2 expression [5, 123, 133, 172-175]. One reason for the upregulation of NSUN2 expression is that the SUMOylation of NSUN2 on the basis of SUMO-2/3 promotes its stability [5]. In addition, studies have shown that the transcription factor E2F1 can activate NSUN2 expression via the AMPK pathway in a peritoneal high-fat environment. Increased NSUN2 regulates ORAI2 mRNA stability through m5C modification via YBX1 recognition, thereby promoting ORAI2 expression and accelerating peritoneal metastasis via PI3K-AKT signalling in GC [173]. Notably, in addition to being recognized by NSUN2, lncRNAs can also reversibly regulate NSUN2 expression and enrichment to further exert m5C-based functions in cells. For example, FOXC2-AS1 increases the m5C methylation level of FOXC2 mRNA by recruiting NSUN2, which is further recognized by YBX1 and regulates the proliferation, migration, and invasion of tumour cells [174]. In addition, DIAPH2-AS1 upregulates the expression of NSUN2 by stabilizing the NSUN2 protein and promotes the epitranscriptomic modification of NTN1 mRNA in gastrointestinal cancer cells [172].
4.3.3 Liver cancer
The main type of liver cancer is hepatocellular carcinoma (HCC), which is a primary malignant tumour originating from liver epithelial tissue or mesenchymal tissue [176]. The overall m5C modification level and the levels of its RMPs, such as NOP2, NSUN2, ALYREF, and YBX1, are greater in HCC tissues than in adjacent tissues. ALYREF expression is significantly increased in HCC, and ALYREF can directly bind to and stabilize the m5C modification site in the 3′-UTR of EGFR mRNA. The subsequent activation of the STAT3 signalling pathway is a critical regulatory mechanism that mediates EMT [177]. YBX1 is highly expressed in HCC and is associated with a poor prognosis. Analysis of RNA-seq and Ribo-seq data has revealed that RNF115 is the target of YBX1 in regulating HCC development [178]. Mechanistically, YBX1 binds to the m5C site of the RNF115 mRNA 3′-UTR and interacts with EIF4A1 to bridge the 5′-UTR, promoting mRNA circularization and translation. RNF115, an E3 ligase, subsequently mediates K27 ubiquitination and autophagic degradation of DHODH to suppress ferroptosis. The main classic functions of m5C readers in cancer are stability and nuclear export. Interestingly, a new biological mechanism of YBX1 has been discovered in HCC. As a multitarget kinase inhibitor for Raf kinases, sorafenib has been approved as a first-line treatment for advanced HCC by the Food and Drug Administration (FDA) of the United States [179]. Studies on m5C modification in HCC have revealed the role of NSUN2 and ALYREF in sorafenib resistance. By RNA-seq and RNA-BisSeq, several mRNAs, including GRB2, RNF115, AATF, c-MYC, PKM2, and MALAT1, have been identified as targets with abundant m5C sites [180-182]. The enrichment of these mRNAs induces sorafenib resistance through various pathways, such as Ras signalling, glycolysis, and ferroptosis.
4.3.4 Cholangiocarcinoma
Cholangiocarcinoma (CCA) is a significant contributor to cancer-related mortality, and its incidence is increasing on a global scale [183, 184]. NSUN5 and ALYREF have been found to be upregulated in CCA tissues and cells [120, 185]. A recent study has revealed that upregulated NSUN5 in CCA mediates the enrichment of glutaminase by increasing m5C modification at the cytosine 137 site within the untranslated region of GLS mRNA [186]. Furthermore, GLS enhances cancer progression by impeding copper-induced cell death mechanisms. Copper is an essential trace element, and its homeostasis can impact cell metabolic processes and even confer resistance to chemotherapy [187]. However, a surplus of copper leads to cuproptosis [188]. These findings establish a correlation between m5C modification and cuproptosis in CCA for the first time, shedding light on the underlying molecular mechanisms and indicating a potential therapeutic target for this disease.
4.3.5 Pancreatic cancer
Pancreatic cancer (PC) is one of the most lethal solid malignancies in which NSUN2, YBX1, and ALYREF are overexpressed. Notably, the highest incidence of perineural invasion (PNI) manifests mainly by the invasion of tumour cells into nerve tissue and their subsequent spread and metastasis along nerves [189, 190]. The severity of PNI is associated with severe disease-related pain and poor survival [191]. YBX1 enhances the stability of PNI-associated mRNAs, including EGR1, NTRK1, and SMAD7, through m5C modification [192]. The increased secretion of migration inhibition factor (MIF) and tumour necrosis factor-α (TNF-α) promote invasion. Overall, epigenetic cross-talk between YBX1 and PNI in PC cells has been reported to be involved. YBX1 also affects the stability of caspase-8 mRNA via m5C modification, resulting in increased caspase-8 expression and inhibition of RIPK1/RIPK3/MLKL pathway phosphorylation in PC [193]. Overexpressed ALYREF might be a novel target for modulating pancreatic ductal adenocarcinoma (PDAC) metabolic vulnerability and immune surveillance [194]. Investigations involving the JASPAR database and RNA-seq data have revealed that ALYREF specifically recognizes and stabilizes JunD mRNA, whose protein serves as a transcription factor of SLC7A5. As SLC7A5 is a key transporter of large neutral amino acids (LNAAs), the overexpression of SLC7A5 in tumour cells depletes amino acids in the TME and restricts the function of CD8+ T cells [195]. In addition, the aberrant m5C modification mediated by NSUN2 in PC is associated with the upregulated expression of TIAM2 mRNA, which promotes EMT and the likelihood of cancer cell migration [196].
4.3.6 Colorectal cancer
In colorectal cancer (CRC), tissue immunohistochemistry has demonstrated an elevated level of m5C modification in tumour tissues compared with adjacent normal tissues. The m5C methyltransferases NSUN2, NSUN4, NSUN5 and the reader protein ALYREF exhibit significantly elevated expression and exert oncogenic functions [129, 197-200]. By RNA-Seq and RNA-BisSeq, NSUN2 and YBX1 have been identified as "writers" and "readers" of ENO1 and SKIL mRNAs in CRC cells. ENO1, the core catalytic enzyme of glycolysis, ultimately reprogrammes glucose metabolism and increases lactate production in an m5C-dependent manner. Interestingly, lactate accumulation in tumour cells, in turn, activates NSUN2 transcription via histone H3K18 lactylation (H3K18la) and induces NSUN2 lactylation at residue Lys356 (K356), which is essential for target RNA capture. The positive-feedback loop of the NSUN2/YBX1/m5C-ENO1 axis connects epigenetic remodelling and metabolic reprogramming [197]. However, the elevated stability of SKIL mRNA ultimately increases transcriptional coactivator with PDZ-binding motif (TAZ) activation [198]. As the first barrier of the body's defense, innate immunity plays a key role in tumour immune surveillance and anti-tumour response, in which type I interferon (IFN-I) is an important mediator with significant antiviral and anti-tumour functions [201-203]. cGAS-STING signaling is a cytosolic DNA-sensing pathway that activates the expression of IFN-I [204, 205]. In colon adenocarcinoma (COAD), GPX4 has emerged as the vital enzyme to prevent lipid peroxidation and maintain cellular redox homeostasis [206, 207]. And NSUN5-mediated m5C modification on GPX4 mRNA facilitated anticancer immunity via activation of cGAS-STING signaling by maintaining redox homeostasis [208, 209]. Accumulating evidence has demonstrated the pivotal role of STING in the antitumour immune response, and the current receptor agonist exhibits potent antineoplastic activity in an immunocompetent mouse model of colon cancer [210]. Therefore, m5C-regulated STING activation holds great potential for therapeutic intervention in cancer immunotherapy. Correlation analysis using the TCGA database and an RIP assay has revealed the direct binding of NSUN4 to NXPH4 mRNA. By relying on the m5C-dependent mechanism, NXPH4 mRNA can avoid degradation by RNautophagy. Furthermore, the competitive binding of the NXPH4 protein with PHD4 impedes HIF1A degradation and activates the HIF signalling pathway. Collectively, these results underscore a new regulatory pathway in which m5C-based NXPH4 plays a pivotal role in driving CRC progression [200]. ALYREF is highly expressed in CRC tissues and predictive of a poor patient prognosis. Integrated analysis of the RIP-BisSeq and transcriptome profiles has revealed RPS6KB2 and RPTOR mRNAs as its downstream effectors. Additionally, ALYREF promotes tumour growth and migration by recruiting ELAVL1 to facilitate the nuclear export of these two transcripts [211].
4.4 Urinary system cancers
4.4.1 Renal cell carcinoma
Clear cell renal cell carcinoma (ccRCC) patients are usually diagnosed at late stages [212]. Therefore, it is imperative to find new strategies for ccRCC therapy. Excitingly, the overexpression of m5C RMPs, NOP2, and YBX1 has provided key insights into the treatment of solid ccRCC tumours [213]. Several analyses, including analyses of TCGA transcriptome profiles, RNA-seq data, and BisSeq data, have revealed that APOL1, a participant in lipid transport and metabolism [214], is a downstream mRNA regulated by NOP2. YBX1 subsequently stabilizes APOL1 mRNA by binding to the m5C site in the 3′-UTR, thus affecting ccRCC cell proliferation, migration, and invasion through the PI3K-Akt pathway. YBX1 also recognizes PEBP1 mRNA via PEBP1P2 recruitment [215]. PEBP1 is a crucial ferroptosis regulator that mediates many cancer-related processes, such as tumour development, metastasis, and the microenvironment [216, 217].
4.4.2 Bladder cancer
m5C is frequently hypermethylated in urothelial carcinoma of the bladder (UCB) and enriched in oncogenic pathways, and NSUN2 and ALYREF have been found to be upregulated in these tissues. Interestingly, the aberrant expression of NSUN2 protein is partially attributed to high levels of m5C methylation of its mRNA [218]. More specifically, ALYREF recognizes the hypermethylated m5C site of NSUN2 mRNA, resulting in NSUN2 upregulation in UCB. BisSeq, RNA-seq, and RIP-seq analyses have revealed that elevated NSUN2 and ALYREF specifically bind to the m5C site in the target TK1 and RABL6 pre-mRNAs, contributing to splicing and stabilization. These results suggest a novel m5C-dependent mechanism of TK1 and RABL6 oncogene expression that enhances the proliferation and invasion of UCB cells. In addition, NSUN2 regulates the m5C site in the 3′-UTR of HDGF mRNA, and YBX1 controls its stability through the indole ring of W65 in its cold shock domain [47]. As a well-known oncogene, HDGF is positively associated with aggressive UCB [219]. HIF-1α induces transcriptional activation of ALYREF, which binds to m5C sites in the 3′-UTR of PKM2 mRNA and enhances its stability [220]. Hence, PKM2, a key enzyme in glycolysis, is upregulated and promotes the proliferation of cancer cells [221]. These findings suggest that NSUN2, YBX1, and ALYREF play oncogenic roles in bladder cancer and participate in the complex regulatory network, providing new insights into the mechanisms of m5C modification in cancer.
4.4.3 Prostate cancer
The m5C RNMTs NSUN2 and NSUN5 are expressed at higher levels in prostate cancer (PCa) tissues than in adjacent tissues. ACC1 is the first rate-limiting enzyme for fatty acid synthesis [222]. Interestingly, phosphorylated NSUN5 increases the m5C modification on ACC1 mRNA in PCa and enhances its stability and nuclear export with the recognition of ALYREF, thereby mediating CDK13-induced lipid accumulation and synthesis to promote PCa growth [223]. These findings indicate that a previously unrecognized m5C-based CDK13-NSUN5-ACC1 axis mediates fatty acid synthesis and lipid accumulation in PCa cells. Lipid metabolism is an extremely important metabolic change in the TME of PCa [224, 225]. NSUN2 expression is also upregulated in PCa and is associated with a poor prognosis [226]. Epitranscriptome assays with RNA-BisSeq analysis have revealed that the 5′-end regions of AR mRNA are modified by NSUN2 and stabilized by an m5C-YBX1-dependent mechanism, which influences several AR variants, including AR-V7. AR is one of the most crucial therapeutic targets in PCa [227, 228]. Since 2012, several new AR inhibitors, such as enzalutamide, abiraterone, and apalutamide, have been approved to treat castration-resistant PCa [229]. However, stimulation of AR variants by AR inhibitors could induce drug resistance because of self-activation without androgen binding [230]. The positive feedback between NSUN2 and AR provides novel evidence that m5C modification clusters exist in PCa and explain cancer progression and the occurrence of castration-resistant PCa.
4.5 Gynaecological cancers
4.5.1 Cervical cancer
Radiotherapy is the main treatment for advanced cervical cancer (CC) [231]. The level of m5C modification is greater in patients with radioresistance, which is related to overexpression of NSUN6 m5C protein and associated with a poor prognosis. Integration of MeRIP-seq and mRNA-seq analysis has revealed that NDGR1 is a downstream target mRNA of NSUN6 and that its stability is increased by specific binding to the m5C reader ALYREF. Abnormal overexpression of NDGR1 promotes homologous recombination (HR)-mediated DNA damage repair (DDR) by recruiting and stabilizing the HR-related protein RDA51, which leads to radiotherapy resistance in CC [232]. Additionally, NSUN2 is also upregulated in CC, increases m5C modification on LRRC8A and KRT13 mRNA, and promotes tumour cell migration and invasion via the YBX1 reader [233, 234]. When cancer cells swell, volume-regulated anion channels (VRACs) are activated [235, 236]. LRRC8A has recently been identified as an essential component of VRACs that can promote the proliferation and migration of cancer cells in CC. Although the role of KRT13 is different in distinct cancers depending on the context [237, 238], research has revealed that the NSUN2-YBX1-KRT13 pathway stimulates CC cell migration and invasion.
4.5.2 Endometrial cancer
Endometrial cancer (EC), the incidence of which has increased by more than 50% during the past two decades, is the most common cancer within the female reproductive system in developed countries [171]. NSUN2 and YBX1 are significantly overexpressed in EC [239]. BisSeq analysis of mRNAs derived from HEC-1B cells has revealed enrichment of ferroptosis-related pathways among differentially methylated genes. Furthermore, NSUN2 promotes SLC7A11 mRNA stability via the recognition role of YBX1, which resists the ferroptosis pathway of tumour cells to promote survival. These results provide new insight into the mechanisms of m5C-based ferroptosis regulation and suggest a promising treatment strategy for EC patients.
4.5.3 Ovarian cancer
Ovarian cancer (OC) has the highest death rate and the worst prognosis of all gynaecological tumours [240], and cytoreductive surgery combined with chemotherapy remains the gold standard of treatment [241]. However, chemotherapy resistance followed by intraperitoneal dissemination still leads to unpredictable deaths. YBX1, an m5C reader, is highly expressed and maintains the stability of various mRNAs, including CDH3, E2F5, YY1, and RCC2, by recognizing their m5C sites, which ultimately leads to drug resistance in cancer cells [242, 243]. Notably, CHD3, an important member of the chromodomain helicase DNA-binding protein (CHD) family, which is involved in regulating chromatin remodelling [244, 245], is a key protein in the response of YBX1 to stress induced by platinum-based drugs. Specifically, highly expressed CHD3 promotes chromatin opening and further enhances HR repair by HR-related proteins such as BRCA1 and RAD50. Platinum resistance is the primary barrier affecting the prognosis of OC patients; hence, the working model of the m5C-CDH3-chromatin accessibility-HR repair axis proposed by these researchers is important for developing therapies that can reverse platinum resistance. In addition, the presence of NOP2 and NSUN2 in OC is associated with the hypermethylation of RAPGEF4 and E2F1 mRNA, respectively, leading to uncontrolled proliferation, migration, and invasion of tumour cells [246, 247].
4.5.4 Breast cancer
Breast cancer (BC) poses a significant threat to women's health because of its intricate pathogenesis and diverse clinical manifestations [248, 249]. Notably, the absolute number of BC cases is increasing in many developing countries due to population growth and the adoption of Western lifestyles [250]. Studies show that most m5C RMPs are significantly dysregulated in BC tissue, and regulate tumorigenesis, progression, prognosis, drug resistance and immune landscape [251]. The malignant phenotype of BC is partially promoted by the overexpression of NSUN2 and YBX1. Through a combination of RNA-Seq and RNA-BisSeq, HGH1 has been identified as a target RNA of NSUN2 [252]. YBX1 synergistically regulates the expression of HGH1 in an m5C-dependent manner by increasing its RNA stability and overall protein synthesis efficiency. The role of HGH1 in human physiology and pathology has rarely been reported. These results preliminarily clarify the biological role that HGH1 might play in the progression of BC. The findings of the m5C mechanism in HGH1 mRNA also reveal a regulatory pathway from posttranscriptional modification to protein translation. Additionally, YBX1, which is stably mediated by SAT1, recognizes the m5C modification site of mTOR mRNA and significantly inhibits autophagy through this gatekeeper of the mTOR signalling pathway in triple-negative BC (TNBC) [253]. The involvement of m5C modification in TNBC, the most aggressive subtype with the poorest prognosis, reveal the complicated interaction between autophagy and tumour progression.
5. Clinical Implications of m5C Modification in Cancer
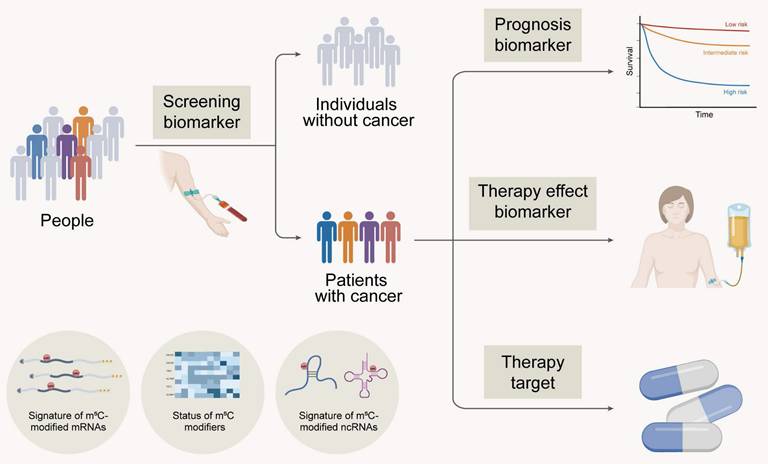
Recently, the profiles and signatures of m5C in RNA, including the expression and mutation of m5C proteins and the m5C modification levels of mRNA and ncRNA, are closely related to the clinical characteristics of patients with tumours. These findings suggest that m5C, as a potential biomarker and therapeutic target, is expected to be applied in clinical practice to benefit cancer patients (Figure 5).
5.1 m5C as a biological marker
Technical advances over the past two decades, especially the unprecedented progression of next-generation sequencing (NGS) technology, have enabled robust diagnosis and detection of cancer in biological samples. In addition to tissue biopsy, liquid-based biopsy assays have been proposed, with a focus on biomarkers, circulating tumour cells (CTCs) [254], circulating tumour DNA (ctDNA) [255], tumour-induced extracellular vehicles (EVs) [256], and other components in body fluids such as blood, urine, and saliva. The level of m5C modification and the status of m5C RMPs are associated with tumorigenesis. Yin et al. reported that the m5C level in peripheral blood immune cells was significantly increased in patients with colorectal cancer and that the degree of m5C modification was positively correlated with tumour progression and metastasis. Therefore, m5C methylation in peripheral blood immune cells is a promising biomarker for noninvasive diagnosis [257].
The clinical and pathological characteristics of tumours, such as stage, pathological type, and treatment sensitivity, determine the prognosis of patients. Increasing evidence has shown that m5C plays an important role in cancer [258]. Therefore, m5C-related features have become a powerful tool for predicting patient prognosis. Huang et al. constructed a survival prediction model for patients with TNBC on the basis of the mRNA expression profiles of NSUN6 and NSUN2 in the TCGA database. The risk score of each patient was calculated using the following formula: risk score = -0.5714 × NSUN6 + 0.024 × NSUN2, where NSUN6 is a protective factor, and NSUN2 is a risk factor. The prediction model shows good performance in evaluating the overall survival (OS) of patients in the public database [259]. According to the TCGA data, genetic alterations in endogenous m5C RMPs were observed in 236 out of 297 CC patients (79%) [260]. This high prevalence underscores the translational potential of these alterations as promising diagnostic biomarkers and therapeutic targets. Based on consistent clustering map of 13 m5C RMPs, upregulation of NSUN2, NSUN3, NSUN6, and TET2, coupled with the downregulation of NSUN5 and ALYREF, is associated with poor survival outcomes of CC patients. Besides, a 4-gene m5C signature comprising FNDC3A, VEGFA, OPN3, and CPE has also demonstrated remarkable 1-year, 3-years and 5-years prognostic capabilities [261]. This refined understanding of m5C RMPs and gene signatures enables the development of a novel molecular diagnostic test, facilitating prognostic assessment and the identification of potential therapeutic targets for CC patients. By integrating mRNA expression data from TCGA, GEO, and real-world cohorts, Liu et al. successfully identified 6 candidate m5C-related genes (SOCS2, LCAT, FTCD, KRT17, PBK, and CBX2) and constructed an m5C scoring model that can be used to effectively predict the prognosis of patients. Survival analysis in the real-world cohort (2-△△CT-based risk score) revealed that the prognostic risk score model was a strong independent prognostic factor.
Treatment resistance in cancer is challenging for doctors regarding the decision-making process in clinical practice. For example, pancreatic ductal adenocarcinoma is the most aggressive malignant tumour of the digestive tract and is highly resistant to treatment. Duo et al. used unsupervised consensus clustering analyses, LASSO, and multivariate Cox regression analysis to construct an m5C scoring signature (m5C score). They reported that the m5C score was associated with the activation of cancer-related pathways, including the Ras, MAPK, and PI3K pathways. Therefore, the sensitivity of patients to pathway-specific inhibitors of PARP, EGFR, AKT, HER2, and mTOR could be evaluated to guide the use of targeted drugs [262].
Clinical implications of m5C modification in cancer. Created in BioRender. Mao, Z. (2025) https://BioRender.com/49digcl.
5.2 m5C as a therapeutic target
Tumour-targeted therapy, also known as molecular-targeted drug therapy, refers to drugs or biological products that inhibit tumour growth and development in local tumour tissue. Such approaches can reduce the toxic effects on normal cells by inhibiting the key signalling pathways involved in tumour initiation and progression and provide more precise and effective strategies. NSUN2 expression is significantly increased in CRC and plays a carcinogenic role. Chen et al. identified a biologically active small-molecule inhibitor in the ChemDiv library that could effectively inhibit NSUN2 expression. The NSUN2 inhibitor NSUN2-i4 significantly enhances the efficacy of PD-1 against colorectal cancer without causing significant toxicity, indicating that NSUN2 is has promise as a target for cancer immunotherapy combined with an immune checkpoint inhibitor (ICI) [197]. Research has revealed that NSUN2 is upregulated in AML and that the inhibition of NSUN2 prevents AML progression in vivo in xenograft experiments [263]. These results indicate that targeting NSUN2 may offer new strategies for treating AML. SU056 is an azodiamidazole-like small molecule that efficiently inhibits the function of the YBX1 protein. Recent studies have shown that targeting YBX1 is expected to reverse platinum resistance in ovarian cancer [242]. 5-Fluorouracil (5-FU) is a first-line chemotherapeutic agent for advanced GC [264]. YBX1 is significantly upregulated in 5-FU-resistant GC cell lines and patient tissues, and YBX1 knockdown increases apoptosis in resistant cells treated with 5-FU [265]. These findings establish YBX1 as a key regulator of autophagy and 5-FU resistance in GC and highlight its potential as a novel therapeutic target for overcoming 5-FU resistance.
6. Conclusion and Future Perspectives
Lifestyle changes, increased access to early screening, and improved treatment continue to reduce cancer-related mortality. However, the incidence of malignant tumours such as those of breast, prostate, and endometrial cancer continues to increase annually. The morbidity of cervical and colorectal cancer tends to be greater in younger patients, which causes serious economic and social burdens around the world. Recently, with advances in technology, in-depth research in the field of epitranscriptomics has revealed the critical role of m5C RNA modification in regulating many cellular pathways [122, 192, 266-268]. However, its potential functions in cancer have not been fully explored. To date, m5C modification is common in rRNA [269], but evidence that rRNA m5C modification regulates reprogramming in cancer is currently lacking. Whether there are more m5C regulatory proteins requires further analysis and demonstration [270, 271]. Moreover, whether RMPs exhibit selectivity or complementarity for the m5C modification sites of RNA is worth further investigation. In particular, why does m5C modification occur in specific mRNAs during cancer progression? We postulate that the m5C modification has a stoichiometric effect. In general, numerous RNAs undergo chemical modification to varying degrees, resulting in a dynamic and reversible equilibrium process. However, during the initiation, progression, and treatment of malignant tumours, dysregulation of RMPs elevates RNA modification levels beyond a critical dose threshold. This disruption breaks the dynamic equilibrium state, rendering it irreversible. Although current research cannot systematically explain the substrate specificity of m5C modification, we propose the following three hypotheses: high CG content, high transcriptome abundance, and structural accessibility. First, specific RNAs possess a high CG content, making them more readily recognizable by RNMTs. Additionally, some RNAs constitute a relatively large proportion of the overall transcriptome, consequently increasing the probability of modification events. Furthermore, the structural conformation of these RNAs renders potential m5C sites more exposed, thereby increasing their accessibility to catalytic enzymes. Hence, a more detailed examination of the regulatory patterns of m5C modification in different parts of a single transcript is essential for advancing our understanding of pathophysiological processes. The small molecules NSUN2-i4 and SU056, which are NSUN2 and YBX1 inhibitors, have been demonstrated to enhance the efficacy of immunotherapy and chemotherapy in mouse models. However, the development of drugs that target m5C modification is still a long way off. Owing to the success of mRNA vaccines in the prevention and treatment of infectious diseases, we are interested in the use of mRNA vaccines in the context of cancer immunotherapy [272]. Whether m5C modification could be applied to mRNA vaccine development deserves further consideration. In addition, in almost all types of malignant tumours, the overall level of m5C modification is elevated, which is related to the deposition of writer proteins in tumour cells. Generally, the factors influencing the expression of writers include genomic mutations and environmental changes. For example, persistent high-risk HPV infection can interfere with the expression of RNMPs at the DNA, RNA and protein levels in cervical cancer. Understanding the upstream regulatory elements of RNMPs can provide valuable insights for clarifying the origin of cancer, developing screening methods, and preventing cancer. In summary, the characteristics, mechanisms, and potential application value of m5C modification in cancer need further exploration.
Abbreviations
m5C: 5-methylcytidine
BisSeq: bisulphite sequencing
ncRNA: noncoding RNA
RMPs: RNA-modifying proteins
SAM: S-adenosyl-methionine
CDS: the coding sequence
MeRIP-seq: methylated RNA immunoprecipitation sequencing
NSUN: NOL1/NOP2/SUN
DNMT: DNA methyltransferase
ALKBH1: alkB homologue 1
ALYREF: Aly/REF export factor
YBX1: Y-box binding protein 1
RNMTs: RNA methyltransferases
TRD: target recognition domain
TRED: target recognition extension domain
PARPis: PARP inhibitors
TC-HR: transcription coupled-homologous recombination
Alt-NHEJ: alternative nonhomologous end joining
DSB: double-strand break
VL: variable loop
SSU: small subunit
LSU: large subunit
CRD: cysteine-rich domain
DSBH: double-stranded β-helix
ESC: embryonic stem cell
NRL: nucleotide recognition lid
CSD: cold shock domain
CTD: C-terminal Domain
OS: osteosarcoma
HNSCC: head and neck squamous cell carcinoma
EMT: epithelial-mesenchymal transition
LSCs: leukemia stem cells
BM: bone marrow
ATC: anaplastic thyroid cancer
LPA: lysophosphatidic acid
TAMs: tumour-associated macrophages
RB: retinoblastoma
NSCLC: non-small cell lung cancer
NPC: nasopharyngeal carcinoma
DMFS: distant metastasis-free survival
OS: overall survival
ESCC: esophageal squamous cell carcinoma
L-OHP: oxaliplatin
VLDL: low-density lipoprotein
HDL: high-density lipoprotein
GC: gastric cancer
HCC: hepatocellular carcinoma
FDA: the Food and Drug Administration
CCA: cholangiocarcinoma
PC: pancreatic cancer
PNI: perineural invasion
MIF: migration inhibition factor
TNF-α: tumour necrosis factor-α
PDAC: pancreatic ductal adenocarcinoma
LNAAs: large neutral amino acids
CRC: colorectal cancer
TAZ: PDZ-binding motif
IFN-I: type I interferon
COAD: colon adenocarcinoma
ccRCC: clear cell renal cell carcinoma
UCB: urothelial carcinoma of the bladder
PCa: prostate cancer
CC: cervical cancer
HR: homologous recombination
DDR: DNA damage repair
VRAC: volume-regulated anion channel
EC: endometrial cancer
OC: ovarian cancer
CHD: DNA-binding protein
BC: breast cancer
TNBC: triple-negative breast cancer
NGS: next-generation sequencing
CTCs: circulating tumour cells
ctDNA: circulating tumour DNA
EVs: extracellular vehicles
ICB: immune checkpoint inhibitor
Acknowledgements
We express our sincere thanks to all authors of this study. The study was supported by The National Natural Science Foundation of China (No. U20A20368), National Key Research and Development Program of China (No. 2021YFC2701202) for Y. Z and Natural Science Foundation of Hunan Province (No. 2025JJ70423) for Y. T. We also thank Biorender.com for creating all the figures, which have obtained publication licensing rights in the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
2. Fujii M, Sekine S, Sato T. Decoding the basis of histological variation in human cancer. Nat Rev Cancer. 2024;24:141-58
3. Song H, Liu D, Dong S, Zeng L, Wu Z, Zhao P. et al. Epitranscriptomics and epiproteomics in cancer drug resistance: therapeutic implications. Signal Transduct Target Ther. 2020;5:193
4. Zuo S, Li L, Wen X, Gu X, Zhuang A, Li R. et al. NSUN2-mediated m(5) C RNA methylation dictates retinoblastoma progression through promoting PFAS mRNA stability and expression. Clin Transl Med. 2023;13:e1273
5. Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B. et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021;12:842
6. Lin S, Kuang M. RNA modification-mediated mRNA translation regulation in liver cancer: mechanisms and clinical perspectives. Nat Rev Gastroenterol Hepatol. 2024;21:267-81
7. Nombela P, Miguel-López B, Blanco S. The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer. 2021;20:18
8. Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023-33
9. Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S. et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582-5
10. Yang X, Yang Y, Sun BF, Chen YS, Xu JW, Lai WY. et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606-25
11. Tang J, Zhou C, Ye F, Zuo S, Zhou M, Lu L. et al. RNA methylation homeostasis in ocular diseases: All eyes on Me. Prog Retin Eye Res. 2025;105:101335
12. Xhemalçe B, Miller KM, Gromak N. Epitranscriptome in action: RNA modifications in the DNA damage response. Mol Cell. 2024;84:3610-26
13. Lu Y, Yang L, Feng Q, Liu Y, Sun X, Liu D. et al. RNA 5-Methylcytosine Modification: Regulatory Molecules, Biological Functions, and Human Diseases. Genomics Proteomics Bioinformatics. 2024;22:qzae063
14. Berggren KA, Schwartz RE, Kleiner RE, Ploss A. The impact of epitranscriptomic modifications on liver disease. Trends Endocrinol Metab. 2024;35:331-46
15. Kong Y, Yu J, Ge S, Fan X. Novel insight into RNA modifications in tumor immunity: Promising targets to prevent tumor immune escape. Innovation (Camb). 2023;4:100452
16. Li Y, Jin H, Li Q, Shi L, Mao Y, Zhao L. The role of RNA methylation in tumor immunity and its potential in immunotherapy. Mol Cancer. 2024;23:130
17. Sun H, Li K, Liu C, Yi C. Regulation and functions of non-m(6)A mRNA modifications. Nat Rev Mol Cell Biol. 2023;24:714-31
18. Liu WW, Zheng SQ, Li T, Fei YF, Wang C, Zhang S. et al. RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy. Signal Transduct Target Ther. 2024;9:70
19. Zhang L, Yuan J, Yao S, Wen G, An J, Jin H. et al. Role of m5C methylation in digestive system tumors (Review). Mol Med Rep. 2025;31:142
20. Song H, Zhang J, Liu B, Xu J, Cai B, Yang H. et al. Biological roles of RNA m(5)C modification and its implications in Cancer immunotherapy. Biomark Res. 2022;10:15
21. Song J, Yi C. Reading Chemical Modifications in the Transcriptome. J Mol Biol. 2020;432:1824-39
22. Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013;9:e1003602
23. Chen YS, Yang WL, Zhao YL, Yang YG. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12:e1639
24. Yang W, Zhao Y, Yang Y. Dynamic RNA methylation modifications and their regulatory role in mammalian development and diseases. Sci China Life Sci. 2024;67:2084-104
25. Lu L, Zhang X, Zhou Y, Shi Z, Xie X, Zhang X. et al. Base-resolution m(5)C profiling across the mammalian transcriptome by bisulfite-free enzyme-assisted chemical labeling approach. Mol Cell. 2024;84:2984-3000.e8
26. Cui X, Liang Z, Shen L, Zhang Q, Bao S, Geng Y. et al. 5-Methylcytosine RNA Methylation in Arabidopsis Thaliana. Mol Plant. 2017;10:1387-99
27. Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009;37:e12
28. Dai Q, Ye C, Irkliyenko I, Wang Y, Sun HL, Gao Y. et al. Ultrafast bisulfite sequencing detection of 5-methylcytosine in DNA and RNA. Nat Biotechnol. 2024;42:1559-70
29. Chen D, Gu X, Nurzat Y, Xu L, Li X, Wu L. et al. Writers, readers, and erasers RNA modifications and drug resistance in cancer. Mol Cancer. 2024;23:178
30. Li M, Tao Z, Zhao Y, Li L, Zheng J, Li Z. et al. 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J Transl Med. 2022;20:214
31. Sharma S, Yang J, Watzinger P, Kötter P, Entian KD. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41:9062-76
32. Brzezicha B, Schmidt M, Makalowska I, Jarmolowski A, Pienkowska J, Szweykowska-Kulinska Z. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA). Nucleic Acids Res. 2006;34:6034-43
33. Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. 2016;12:546-51
34. Metodiev MD, Spåhr H, Loguercio Polosa P, Meharg C, Becker C, Altmueller J. et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014;10:e1004110
35. Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E. et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun. 2015;6:6158
36. Liu RJ, Long T, Li J, Li H, Wang ED. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017;45:6684-97
37. Harris T, Marquez B, Suarez S, Schimenti J. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol Reprod. 2007;77:376-82
38. Li H, Liu H, Zhu D, Dou C, Gang B, Zhang M. et al. Biological function molecular pathways and druggability of DNMT2/TRDMT1. Pharmacol Res. 2024;205:107222
39. Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X. et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395-8
40. Zhang X, Zhang Y, Wang C, Wang X. TET (Ten-eleven translocation) family proteins: structure, biological functions and applications. Signal Transduct Target Ther. 2023;8:297
41. Shen H, Ontiveros RJ, Owens MC, Liu MY, Ghanty U, Kohli RM. et al. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J Biol Chem. 2021;296:100087
42. Arguello AE, Li A, Sun X, Eggert TW, Mairhofer E, Kleiner RE. Reactivity-dependent profiling of RNA 5-methylcytidine dioxygenases. Nat Commun. 2022;13:4176
43. Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, Suzuki T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017;45:7401-15
44. Zhao Y, Xing C, Peng H. ALYREF (Aly/REF export factor): A potential biomarker for predicting cancer occurrence and therapeutic efficacy. Life Sci. 2024;338:122372
45. Xue C, Gu X, Zheng Q, Shi Q, Yuan X, Su Y. et al. ALYREF mediates RNA m(5)C modification to promote hepatocellular carcinoma progression. Signal Transduct Target Ther. 2023;8:130
46. Dinh NTM, Nguyen TM, Park MK, Lee CH. Y-Box Binding Protein 1: Unraveling the Multifaceted Role in Cancer Development and Therapeutic Potential. Int J Mol Sci. 2024;25:717
47. Chen X, Li A, Sun BF, Yang Y, Han YN, Yuan X. et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978-90
48. Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL. et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol Cell. 2019;75:1188-202.e11
49. Motorin Y, Lyko F, Helm M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415-30
50. Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453-63
51. Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V. et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:856-63
52. Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. Embo j. 2014;33:2020-39
53. Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255-61
54. Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. Rna. 1999;5:1105-18
55. Muhammad N, Hussain SI, Rehman ZU, Khan SA, Jan S, Khan N. et al. Autosomal recessive variants c.953A>C and c.97-1G>C in NSUN2 causing intellectual disability: a molecular dynamics simulation study of loss-of-function mechanisms. Front Neurol. 2023;14:1168307
56. Plotkin JB, Robins H, Levine AJ. Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci U S A. 2004;101:12588-91
57. Li P, Huang D. NSUN2-mediated RNA methylation: Molecular mechanisms and clinical relevance in cancer. Cell Signal. 2024;123:111375
58. Jiang J, Liu F, Cui D, Xu C, Chi J, Yan T. et al. Novel molecular mechanisms of immune evasion in hepatocellular carcinoma: NSUN2-mediated increase of SOAT2 RNA methylation. Cancer Commun (Lond). 2025 [Epub ahead of print]
59. Qi J, Jiang T, Liu B, Hu Q, Chen J, Ma N. et al. LINC02167 stabilizes KSR1 mRNA in an m(5)C-dependent manner to regulate the ERK/MAPK signaling pathway and promotes colorectal cancer metastasis. J Exp Clin Cancer Res. 2025;44:121
60. Wang Z, Mierxiati A, Zhu W, Li T, Xu H, Wan F. et al. FOXA1-dependent NSUN2 facilitates the advancement of prostate cancer by preserving TRIM28 mRNA stability in a m5C-dependent manner. NPJ Precis Oncol. 2025;9:127
61. Chen T, Xu ZG, Luo J, Manne RK, Wang Z, Hsu CC. et al. NSUN2 is a glucose sensor suppressing cGAS/STING to maintain tumorigenesis and immunotherapy resistance. Cell Metab. 2023;35:1782-98.e8
62. Liao H, Gaur A, McConie H, Shekar A, Wang K, Chang JT. et al. Human NOP2/NSUN1 regulates ribosome biogenesis through non-catalytic complex formation with box C/D snoRNPs. Nucleic Acids Res. 2022;50:10695-716
63. Delaunay S, Pascual G, Feng B, Klann K, Behm M, Hotz-Wagenblatt A. et al. Mitochondrial RNA modifications shape metabolic plasticity in metastasis. Nature. 2022;607:593-603
64. Cámara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B. et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527-39
65. Janin M, Ortiz-Barahona V, de Moura MC, Martínez-Cardús A, Llinàs-Arias P, Soler M. et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053-74
66. Li J, Li H, Long T, Dong H, Wang ED, Liu RJ. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019;47:2041-55
67. Dev RR, Ganji R, Singh SP, Mahalingam S, Banerjee S, Khosla S. Cytosine methylation by DNMT2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem J. 2017;474:2009-26
68. Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S. et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900-5
69. Shanmugam R, Aklujkar M, Schäfer M, Reinhardt R, Nickel O, Reuter G. et al. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res. 2014;42:6487-96
70. Burgess AL, David R, Searle IR. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 2015;15:199
71. Huang ZX, Li J, Xiong QP, Li H, Wang ED, Liu RJ. Position 34 of tRNA is a discriminative element for m5C38 modification by human DNMT2. Nucleic Acids Res. 2021;49:13045-61
72. Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G. et al. Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. Rna. 2008;14:1663-70
73. Li H, Zhu D, Yang Y, Ma Y, Chen Y, Xue P. et al. Determinants of DNMT2/TRDMT1 preference for substrates tRNA and DNA during the evolution. RNA Biol. 2023;20:875-92
74. Becker M, Müller S, Nellen W, Jurkowski TP, Jeltsch A, Ehrenhofer-Murray AE. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 2012;40:11648-58
75. Li H, Zhu D, Wu J, Ma Y, Cai C, Chen Y. et al. New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1. RNA Biol. 2021;18:2531-45
76. Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G. et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. Embo j. 2015;34:2350-62
77. Chen H, Yang H, Zhu X, Yadav T, Ouyang J, Truesdell SS. et al. m(5)C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat Commun. 2020;11:2834
78. Yang H, Lachtara EM, Ran X, Hopkins J, Patel PS, Zhu X. et al. The RNA m5C modification in R-loops as an off switch of Alt-NHEJ. Nat Commun. 2023;14:6114
79. Haag S, Warda AS, Kretschmer J, Günnigmann MA, Höbartner C, Bohnsack MT. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. Rna. 2015;21:1532-43
80. Huang W, Lan MD, Qi CB, Zheng SJ, Wei SZ, Yuan BF. et al. Formation and determination of the oxidation products of 5-methylcytosine in RNA. Chem Sci. 2016;7:5495-502
81. Xue C, Zhao Y, Li L. Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer. Biomark Res. 2020;8:43
82. Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M. et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545-55
83. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-5
84. Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760-4
85. Lan J, Rajan N, Bizet M, Penning A, Singh NK, Guallar D. et al. Functional role of Tet-mediated RNA hydroxymethylcytosine in mouse ES cells and during differentiation. Nat Commun. 2020;11:4956
86. Shen Q, Zhang Q, Shi Y, Shi Q, Jiang Y, Gu Y. et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature. 2018;554:123-7
87. Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R. et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282-5
88. He C, Sidoli S, Warneford-Thomson R, Tatomer DC, Wilusz JE, Garcia BA. et al. High-Resolution Mapping of RNA-Binding Regions in the Nuclear Proteome of Embryonic Stem Cells. Mol Cell. 2016;64:416-30
89. Yang H, Wang Y, Xiang Y, Yadav T, Ouyang J, Phoon L. et al. FMRP promotes transcription-coupled homologous recombination via facilitating TET1-mediated m5C RNA modification demethylation. Proc Natl Acad Sci U S A. 2022;119:e2116251119
90. Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Massé A. et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289-301
91. Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS. et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839-43
92. Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M. et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838-42
93. Li Y, Xue M, Deng X, Dong L, Nguyen LXT, Ren L. et al. TET2-mediated mRNA demethylation regulates leukemia stem cell homing and self-renewal. Cell Stem Cell. 2023;30:1072-90.e10
94. Kosmider O, Delabesse E, de Mas VM, Cornillet-Lefebvre P, Blanchet O, Delmer A. et al. TET2 mutations in secondary acute myeloid leukemias: a French retrospective study. Haematologica. 2011;96:1059-63
95. Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE. et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509-18
96. Zou Z, Dou X, Li Y, Zhang Z, Wang J, Gao B. et al. RNA m(5)C oxidation by TET2 regulates chromatin state and leukaemogenesis. Nature. 2024;634:986-94
97. Zhang J, Tan P, Guo L, Gong J, Ma J, Li J. et al. p53-dependent autophagic degradation of TET2 modulates cancer therapeutic resistance. Oncogene. 2019;38:1905-19
98. Puig I, Tenbaum SP, Chicote I, Arqués O, Martínez-Quintanilla J, Cuesta-Borrás E. et al. TET2 controls chemoresistant slow-cycling cancer cell survival and tumor recurrence. J Clin Invest. 2018;128:3887-905
99. Pan W, Zhu S, Qu K, Meeth K, Cheng J, He K. et al. The DNA Methylcytosine Dioxygenase Tet2 Sustains Immunosuppressive Function of Tumor-Infiltrating Myeloid Cells to Promote Melanoma Progression. Immunity. 2017;47:284-97.e5
100. Zhang Y, Wang C. Demethyltransferase AlkBH1 substrate diversity and relationship to human diseases. Mol Biol Rep. 2021;48:4747-56
101. Tsujikawa K, Koike K, Kitae K, Shinkawa A, Arima H, Suzuki T. et al. Expression and sub-cellular localization of human ABH family molecules. J Cell Mol Med. 2007;11:1105-16
102. Westbye MP, Feyzi E, Aas PA, Vågbø CB, Talstad VA, Kavli B. et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. 2008;283:25046-56
103. Zhang M, Yang S, Nelakanti R, Zhao W, Liu G, Li Z. et al. Mammalian ALKBH1 serves as an N(6)-mA demethylase of unpairing DNA. Cell Res. 2020;30:197-210
104. Zhang C, Li J, Wang L, Yang P, Luo X. ALKBH1 knockdown promotes the growth, migration and invasion of HTR-8/SVneo cells through regulating the m5C modification PSMD14. Sci Rep. 2025;15:7345
105. Shi M, Zhang H, Wu X, He Z, Wang L, Yin S. et al. ALYREF mainly binds to the 5' and the 3' regions of the mRNA in vivo. Nucleic Acids Res. 2017;45:9640-53
106. Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 2010;24:2043-53
107. Tong X, Xiang Y, Hu Y, Hu Y, Li H, Wang H. et al. NSUN2 Promotes Tumor Progression and Regulates Immune Infiltration in Nasopharyngeal Carcinoma. Front Oncol. 2022;12:788801
108. Hou X, Dong Q, Hao J, Liu M, Ning J, Tao M. et al. NSUN2-mediated m(5)C modification drives alternative splicing reprogramming and promotes multidrug resistance in anaplastic thyroid cancer through the NSUN2/SRSF6/UAP1 signaling axis. Theranostics. 2025;15:2757-77
109. Zhang RK, Li Y, Sun FL, Zhou ZH, Xie YX, Liu WJ. et al. RNA methyltransferase NSUN2-mediated m5C methylation promotes Cr(VI)-induced malignant transformation and lung cancer by accelerating metabolism reprogramming. Environ Int. 2024;192:109055
110. Li Z, Wu X. NSUN4 Facilitates the Activity of Oncogenic Protein CDC20 to Promote NSCLC Development by Mediating m5C Modification of CDC20 mRNA. Thorac Cancer. 2025;16:e70023
111. Mordovkina D, Lyabin DN, Smolin EA, Sogorina EM, Ovchinnikov LP, Eliseeva I. Y-Box Binding Proteins in mRNP Assembly, Translation, and Stability Control. Biomolecules. 2020;10:591
112. Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 protein: functions and regulation. Wiley Interdiscip Rev RNA. 2014;5:95-110
113. Yang XJ, Zhu H, Mu SR, Wei WJ, Yuan X, Wang M. et al. Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA. J Biol Chem. 2019;294:10998-1010
114. Evans MK, Matsui Y, Xu B, Willis C, Loome J, Milburn L. et al. Ybx1 fine-tunes PRC2 activities to control embryonic brain development. Nat Commun. 2020;11:4060
115. Kwon E, Todorova K, Wang J, Horos R, Lee KK, Neel VA. et al. The RNA-binding protein YBX1 regulates epidermal progenitors at a posttranscriptional level. Nat Commun. 2018;9:1734
116. Jayavelu AK, Schnöder TM, Perner F, Herzog C, Meiler A, Krishnamoorthy G. et al. Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature. 2020;588:157-63
117. Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell. 2015;161:790-802
118. Yu T, Zhang Q, Yu SK, Nie FQ, Zhang ML, Wang Q. et al. THOC3 interacts with YBX1 to promote lung squamous cell carcinoma progression through PFKFB4 mRNA modification. Cell Death Dis. 2023;14:475
119. Li R, Li S, Shen L, Li J, Zhang D, Yu J. et al. LINC00618 facilitates growth and metastasis of hepatocellular carcinoma via elevating cholesterol synthesis by promoting NSUN2-mediated SREBP2 m5C modification. Ecotoxicol Environ Saf. 2024;285:117064
120. Wang X, Duan W, Ma Z, Wen H, Mao X, Liu C. ETV4/ALYREF-mediated glycolytic metabolism through PKM2 enhances resistance to ferroptosis and promotes the development of intrahepatic cholangiocarcinoma. Cancer Metab. 2025;13:19
121. Meng L, Wu H, Wu J, Ding P, He J, Li T. et al. Aberrant CircTMEM45A Facilitates Inflammatory Progression of Esophageal Squamous Cell Carcinoma through m5C-Mediated NLRP3 Activation. Cancer Res. 2025;85:2694-713
122. Hu S, Yang M, Xiao K, Yang Z, Cai L, Xie Y. et al. Loss of NSUN6 inhibits osteosarcoma progression by downregulating EEF1A2 expression and activation of Akt/mTOR signaling pathway via m(5)C methylation. Exp Ther Med. 2023;26:457
123. Zhao G, Liu R, Ge L, Qi D, Wu Q, Lin Z. et al. NONO regulates m(5)C modification and alternative splicing of PTEN mRNAs to drive gastric cancer progression. J Exp Clin Cancer Res. 2025;44:81
124. Yang M, Wei R, Zhang S, Hu S, Liang X, Yang Z. et al. NSUN2 promotes osteosarcoma progression by enhancing the stability of FABP5 mRNA via m(5)C methylation. Cell Death Dis. 2023;14:125
125. Huang S, Cao C, Tang D, Liu Y, Zhou W, Liu L. et al. NSUN2 Promotes Head and Neck Squamous Cell Carcinoma Progression by Targeting EMT-Related Gene LAMC2 in an m(5)C-YBX1-Dependent Manner. Biomedicines. 2024;12:2533
126. Wang Y, Wei J, Feng L, Li O, Huang L, Zhou S. et al. Aberrant m5C hypermethylation mediates intrinsic resistance to gefitinib through NSUN2/YBX1/QSOX1 axis in EGFR-mutant non-small-cell lung cancer. Mol Cancer. 2023;22:81
127. Li P, Wang W, Zhou R, Ding Y, Li X. The m(5) C methyltransferase NSUN2 promotes codon-dependent oncogenic translation by stabilising tRNA in anaplastic thyroid cancer. Clin Transl Med. 2023;13:e1466
128. Chen S, Cai D, Zhao Q, Wu J, Zhou X, Xu H. et al. NSUN2-mediated m5C modification of circFAM190B promotes lung cancer progression by inhibiting cellular autophagy. Int J Biol Macromol. 2025;306:141528
129. Hou C, Liu J, Liu J, Yao D, Liang F, Qin C. et al. 5-methylcytosine-mediated upregulation of circular RNA 0102913 augments malignant properties of colorectal cancer cells through a microRNA-571/Rac family small GTPase 2 axis. Gene. 2024;901:148162
130. Doye A, Chaintreuil P, Lagresle-Peyrou C, Batistic L, Marion V, Munro P. et al. RAC2 gain-of-function variants causing inborn error of immunity drive NLRP3 inflammasome activation. J Exp Med. 2024;221:e20231562
131. Wu J, Zhao Q, Chen S, Xu H, Zhang R, Cai D. et al. NSUN4-mediated m5C modification of circERI3 promotes lung cancer development by altering mitochondrial energy metabolism. Cancer Lett. 2024;605:217266
132. Pan A, Xue Y, Ruan X, Dong W, Wang D, Liu Y. et al. m5C modification of LINC00324 promotes angiogenesis in glioma through CBX3/VEGFR2 pathway. Int J Biol Macromol. 2024;257:128409
133. Fang L, Huang H, Lv J, Chen Z, Lu C, Jiang T. et al. m5C-methylated lncRNA NR_033928 promotes gastric cancer proliferation by stabilizing GLS mRNA to promote glutamine metabolism reprogramming. Cell Death Dis. 2023;14:520
134. Sun Z, Xue S, Zhang M, Xu H, Hu X, Chen S. et al. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. 2020;39:6906-19
135. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70
136. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
137. Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31-46
138. Zhao Z, Zhou Y, Lv P, Zhou T, Liu H, Xie Y. et al. NSUN4 mediated RNA 5-methylcytosine promotes the malignant progression of glioma through improving the CDC42 mRNA stabilization. Cancer Lett. 2024;597:217059
139. Xu X, Zhang Y, Zhang J, Zhang X. NSun2 promotes cell migration through methylating autotaxin mRNA. J Biol Chem. 2020;295:18134-47
140. Liu S, Umezu-Goto M, Murph M, Lu Y, Liu W, Zhang F. et al. Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell. 2009;15:539-50
141. Nam SW, Clair T, Campo CK, Lee HY, Liotta LA, Stracke ML. Autotaxin (ATX), a potent tumor motogen, augments invasive and metastatic potential of ras-transformed cells. Oncogene. 2000;19:241-7
142. Zhang P, Rashidi A, Zhao J, Silvers C, Wang H, Castro B. et al. STING agonist-loaded, CD47/PD-L1-targeting nanoparticles potentiate antitumor immunity and radiotherapy for glioblastoma. Nat Commun. 2023;14:1610
143. Wu R, Sun C, Chen X, Yang R, Luan Y, Zhao X. et al. NSUN5/TET2-directed chromatin-associated RNA modification of 5-methylcytosine to 5-hydroxymethylcytosine governs glioma immune evasion. Proc Natl Acad Sci U S A. 2024;121:e2321611121
144. Luo G, Xu W, Chen X, Wang S, Wang J, Dong F. et al. NSUN2-mediated RNA m(5)C modification modulates uveal melanoma cell proliferation and migration. Epigenetics. 2022;17:922-33
145. Chen Y, Jiang Z, Zhang C, Zhang L, Chen H, Xiao N. et al. 5-Methylcytosine transferase NSUN2 drives NRF2-mediated ferroptosis resistance in non-small cell lung cancer. J Biol Chem. 2024;300:106793
146. Yang Y, Cao L, Xu X, Li D, Deng Y, Li L. et al. NSUN2/ALYREF axis-driven m(5)C methylation enhances PD-L1 expression and facilitates immune evasion in non-small-cell lung cancer. Cancer Immunol Immunother. 2025;74:132
147. Yang Y, Fan H, Liu H, Lou X, Xiao N, Zhang C. et al. NOP2 facilitates EZH2-mediated epithelial-mesenchymal transition by enhancing EZH2 mRNA stability via m5C methylation in lung cancer progression. Cell Death Dis. 2024;15:506
148. Lu Z, Liu B, Kong D, Zhou X, Pei D, Liu D. NSUN6 Regulates NM23-H1 Expression in an m5C Manner to Affect Epithelial-Mesenchymal Transition in Lung Cancer. Med Princ Pract. 2024;33:56-65
149. Yang Q, Wang M, Xu J, Yu D, Li Y, Chen Y. et al. LINC02159 promotes non-small cell lung cancer progression via ALYREF/YAP1 signaling. Mol Cancer. 2023;22:122
150. Zhou Z, Yin X, Sun H, Lu J, Li Y, Fan Y. et al. PTBP1 Lactylation Promotes Glioma Stem Cell Maintenance through PFKFB4-Driven Glycolysis. Cancer Res. 2025;85:739-57
151. Liu H, Tang L, Li Y, Xie W, Zhang L, Tang H. et al. Nasopharyngeal carcinoma: current views on the tumor microenvironment's impact on drug resistance and clinical outcomes. Mol Cancer. 2024;23:20
152. Liu Q, Wang H, Chen Z, Xiong J, Huang Y, Zhang S. et al. Global, regional, and national epidemiology of nasopharyngeal carcinoma in middle-aged and elderly patients from 1990 to 2021. Ageing Res Rev. 2025;104:102613
153. Jin Y, Yao J, Fu J, Huang Q, Luo Y, You Y. et al. ALYREF promotes the metastasis of nasopharyngeal carcinoma by increasing the stability of NOTCH1 mRNA. Cell Death Dis. 2024;15:578
154. Deboever N, Jones CM, Yamashita K, Ajani JA, Hofstetter WL. Advances in diagnosis and management of cancer of the esophagus. Bmj. 2024;385:e074962
155. Jiang W, Zhang B, Xu J, Xue L, Wang L. Current status and perspectives of esophageal cancer: a comprehensive review. Cancer Commun (Lond). 2025;45:281-331
156. Liu L, Chen Y, Zhang T, Cui G, Wang W, Zhang G. et al. YBX1 Promotes Esophageal Squamous Cell Carcinoma Progression via m5C-Dependent SMOX mRNA Stabilization. Adv Sci (Weinh). 2024;11:e2302379
157. Su J, Wu G, Ye Y, Zhang J, Zeng L, Huang X. et al. NSUN2-mediated RNA 5-methylcytosine promotes esophageal squamous cell carcinoma progression via LIN28B-dependent GRB2 mRNA stabilization. Oncogene. 2021;40:5814-28
158. Cui Y, Hu Z, Zhang C. RNA Methyltransferase NSUN5 Promotes Esophageal Cancer via 5-Methylcytosine Modification of METTL1. Mol Carcinog. 2025;64:399-409
159. Wang D, Liu G, Meng Y, Chen H, Ye Z, Jing J. The Configuration of GRB2 in Protein Interaction and Signal Transduction. Biomolecules. 2024;14:259
160. Qi YN, Liu Z, Hong LL, Li P, Ling ZQ. Methyltransferase-like proteins in cancer biology and potential therapeutic targeting. J Hematol Oncol. 2023;16:89
161. Fang H, He J, Du D, Wang X, Xu X, Lu L. et al. Deciphering the secret codes in N(7)-methylguanosine modification: Context-dependent function of methyltransferase-like 1 in human diseases. Clin Transl Med. 2025;15:e70240
162. Hoeppner J, Brunner T, Schmoor C, Bronsert P, Kulemann B, Claus R. et al. Perioperative Chemotherapy or Preoperative Chemoradiotherapy in Esophageal Cancer. N Engl J Med. 2025;392:323-35
163. Hu J, Liu Q, Feng B, Lu Y, Chen K. ALYREF-Mediated Regulation of TBL1XR1 and KMT2E Synergistically Upregulates APOC1, Contributing to Oxaliplatin Resistance in Esophageal Cancer. Cancer Res Treat. 2025 [Epub ahead of print]
164. Rouland A, Masson D, Lagrost L, Vergès B, Gautier T, Bouillet B. Role of apolipoprotein C1 in lipoprotein metabolism, atherosclerosis and diabetes: a systematic review. Cardiovasc Diabetol. 2022;21:272
165. Ren L, Yi J, Yang Y, Li W, Zheng X, Liu J. et al. Systematic pan-cancer analysis identifies APOC1 as an immunological biomarker which regulates macrophage polarization and promotes tumor metastasis. Pharmacol Res. 2022;183:106376
166. Jia Y, Zhang B, Zhang C, Kwong DL, Chang Z, Li S. et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Esophageal Squamous Cell Carcinoma. Adv Sci (Weinh). 2023;10:e2204565
167. Wu L, Gu L, Zheng Y, Liu J, Wei Z, Liu F. et al. CircPRKCA facilitates esophageal squamous cell carcinoma metastasis via m5C-dependent CSF2 mRNA stabilization. J Transl Med. 2025;23:385
168. Li CF, Chan TC, Fang FM, Yu SC, Huang HY. PAK1 overexpression promotes myxofibrosarcoma angiogenesis through STAT5B-mediated CSF2 transactivation: clinical and therapeutic relevance of amplification and nuclear entry. Int J Biol Sci. 2023;19:3920-36
169. Yu X, Li S, Ke S, Ye C, Wang Q, Wang H. et al. CSF2 Impairs Nrf2 Signaling through the Akt/Mtor Pathway in the Development of Bladder Cancer. J Cancer. 2024;15:3242-53
170. Yonemitsu K, Pan C, Fujiwara Y, Miyasato Y, Shiota T, Yano H. et al. GM-CSF derived from the inflammatory microenvironment potentially enhanced PD-L1 expression on tumor-associated macrophages in human breast cancer. Sci Rep. 2022;12:12007
171. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-63
172. Li Y, Xia Y, Jiang T, Chen Z, Shen Y, Lin J. et al. Long noncoding RNA DIAPH2-AS1 promotes neural invasion of gastric cancer via stabilizing NSUN2 to enhance the m5C modification of NTN1. Cell Death Dis. 2023;14:260
173. Liu K, Xu P, Lv J, Ge H, Yan Z, Huang S. et al. Peritoneal high-fat environment promotes peritoneal metastasis of gastric cancer cells through activation of NSUN2-mediated ORAI2 m5C modification. Oncogene. 2023;42:1980-93
174. Yan J, Liu J, Huang Z, Huang W, Lv J. FOXC2-AS1 stabilizes FOXC2 mRNA via association with NSUN2 in gastric cancer cells. Hum Cell. 2021;34:1755-64
175. Shen X, Sun H, Shu S, Tang W, Yuan Y, Su H. et al. Suppression of NSUN2 enhances the sensitivity to chemosensitivity and inhibits proliferation by mediating cell apoptosis in gastric cancer. Pathol Res Pract. 2024;253:154986
176. Yang X, Yang C, Zhang S, Geng H, Zhu AX, Bernards R. et al. Precision treatment in advanced hepatocellular carcinoma. Cancer Cell. 2024;42:180-97
177. Nulali J, Zhang K, Long M, Wan Y, Liu Y, Zhang Q. et al. ALYREF-mediated RNA 5-Methylcytosine modification Promotes Hepatocellular Carcinoma Progression Via Stabilizing EGFR mRNA and pSTAT3 activation. Int J Biol Sci. 2024;20:331-46
178. Li O, An K, Wang H, Li X, Wang Y, Huang L. et al. Targeting YBX1-m5C mediates RNF115 mRNA circularisation and translation to enhance vulnerability of ferroptosis in hepatocellular carcinoma. Clin Transl Med. 2025;15:e70270
179. Gordan JD, Kennedy EB, Abou-Alfa GK, Beal E, Finn RS, Gade TP. et al. Systemic Therapy for Advanced Hepatocellular Carcinoma: ASCO Guideline Update. J Clin Oncol. 2024;42:1830-50
180. Zhang H, Zhai X, Liu Y, Xia Z, Xia T, Du G. et al. NOP2-mediated m5C Modification of c-Myc in an EIF3A-Dependent Manner to Reprogram Glucose Metabolism and Promote Hepatocellular Carcinoma Progression. Research (Wash D C). 2023;6:0184
181. Song D, An K, Zhai W, Feng L, Xu Y, Sun R. et al. NSUN2-mediated mRNA m(5)C Modification Regulates the Progression of Hepatocellular Carcinoma. Genomics Proteomics Bioinformatics. 2023;21:823-33
182. Qi Q, Zhong R, Huang Y, Tang Y, Zhang XW, Liu C. et al. The RNA M5C methyltransferase NSUN2 promotes progression of hepatocellular carcinoma by enhancing PKM2-mediated glycolysis. Cell Death Dis. 2025;16:82
183. Luo M, Yang J, Zhang K, Sun J, Lu Z, Wang Z. et al. Current advance in comprehensive management of hilar cholangiocarcinoma and navigation in surgery: non-systematic reviews. Int J Surg. 2025;111:2131-47
184. Ye L, Schneider JS, Ben Khaled N, Schirmacher P, Seifert C, Frey L. et al. Combined Hepatocellular-Cholangiocarcinoma: Biology, Diagnosis, and Management. Liver Cancer. 2024;13:6-28
185. Hao Z, Yang H, Zhu W, Yu D, Cao Y, Wu Y. ALYREF Promotes Progression of Intrahepatic Cholangiocarcinoma through Increasing the Level of Isocitrate Dehydrogenase 1 in an m5C-Dependent Manner. Mol Cell Biol. 2025;45:198-211
186. Shu M, Guo K, Huang Y, Wang W. NSUN5 promotes cholangiocarcinoma progression by enhancing GLS mRNA stabilization. J Cancer Res Clin Oncol. 2025;151:117
187. Lutsenko S, Roy S, Tsvetkov P. Mammalian copper homeostasis: physiological roles and molecular mechanisms. Physiol Rev. 2025;105:441-91
188. Tang D, Kroemer G, Kang R. Targeting cuproplasia and cuproptosis in cancer. Nat Rev Clin Oncol. 2024;21:370-88
189. Bapat AA, Hostetter G, Von Hoff DD, Han H. Perineural invasion and associated pain in pancreatic cancer. Nat Rev Cancer. 2011;11:695-707
190. Li J, Kang R, Tang D. Cellular and molecular mechanisms of perineural invasion of pancreatic ductal adenocarcinoma. Cancer Commun (Lond). 2021;41:642-60
191. Yuan H, Zhang Y, Liu F, Wu Y, Huang X, Liu X. et al. Exploring the biological mechanism and clinical value of perineural invasion in pancreatic cancer. Cancer Lett. 2025;613:217515
192. Zheng S, Hu C, Lin Q, Li T, Li G, Tian Q. et al. Extracellular vesicle-packaged PIAT from cancer-associated fibroblasts drives neural remodeling by mediating m5C modification in pancreatic cancer mouse models. Sci Transl Med. 2024;16:eadi0178
193. Yu L, Guo Q, Li Y, Mao M, Liu Z, Li T. et al. CHMP4C promotes pancreatic cancer progression by inhibiting necroptosis via the RIPK1/RIPK3/MLKL pathway. J Adv Res. 2025 [Epub ahead of print]
194. Meng Q, Xie Y, Sun K, He L, Wu H, Zhang Q. et al. ALYREF-JunD-SLC7A5 axis promotes pancreatic ductal adenocarcinoma progression through epitranscriptome-metabolism reprogramming and immune evasion. Cell Death Discov. 2024;10:97
195. Kanai Y. Amino acid transporter LAT1 (SLC7A5) as a molecular target for cancer diagnosis and therapeutics. Pharmacol Ther. 2022;230:107964
196. Zhang G, Liu L, Li J, Chen Y, Wang Y, Zhang Y. et al. NSUN2 stimulates tumor progression via enhancing TIAM2 mRNA stability in pancreatic cancer. Cell Death Discov. 2023;9:219
197. Chen B, Deng Y, Hong Y, Fan L, Zhai X, Hu H. et al. Metabolic Recoding of NSUN2-Mediated m(5)C Modification Promotes the Progression of Colorectal Cancer via the NSUN2/YBX1/m(5)C-ENO1 Positive Feedback Loop. Adv Sci (Weinh). 2024;11:e2309840
198. Zou S, Huang Y, Yang Z, Zhang J, Meng M, Zhang Y. et al. NSUN2 promotes colorectal cancer progression by enhancing SKIL mRNA stabilization. Clin Transl Med. 2024;14:e1621
199. Tong R, Li Y, Wang J, Liu C, Liu Y, Li R. et al. NSUN2 Knockdown Promotes the Ferroptosis of Colorectal Cancer Cells Via m5C Modification of SLC7A11 mRNA. Biochem Genet. 2025 [Epub ahead of print]
200. Yang L, Shi J, Zhong M, Sun P, Zhang X, Lian Z. et al. NXPH4 mediated by m(5)C contributes to the malignant characteristics of colorectal cancer via inhibiting HIF1A degradation. Cell Mol Biol Lett. 2024;29:111
201. Kagan JC. Infection infidelities drive innate immunity. Science. 2023;379:333-5
202. Li Y, Slavik KM, Toyoda HC, Morehouse BR, de Oliveira Mann CC, Elek A. et al. cGLRs are a diverse family of pattern recognition receptors in innate immunity. Cell. 2023;186:3261-76.e20
203. Carpenter S, O'Neill LAJ. From periphery to center stage: 50 years of advancements in innate immunity. Cell. 2024;187:2030-51
204. Zhang Z, Zhang C. Regulation of cGAS-STING signalling and its diversity of cellular outcomes. Nat Rev Immunol. 2025;25:425-44
205. Shen M, Jiang X, Peng Q, Oyang L, Ren Z, Wang J. et al. The cGAS-STING pathway in cancer immunity: mechanisms, challenges, and therapeutic implications. J Hematol Oncol. 2025;18:40
206. Wang X, Chen T, Chen S, Zhang J, Cai L, Liu C. et al. STING aggravates ferroptosis-dependent myocardial ischemia-reperfusion injury by targeting GPX4 for autophagic degradation. Signal Transduct Target Ther. 2025;10:136
207. Liang D, Feng Y, Zandkarimi F, Wang H, Zhang Z, Kim J. et al. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell. 2023;186:2748-64.e22
208. Chen B, Hong Y, Zhai X, Deng Y, Hu H, Tian S. et al. m6A and m5C modification of GPX4 facilitates anticancer immunity via STING activation. Cell Death Dis. 2023;14:809
209. Sun Y, Liu Y, Jiang L, Zhong C. m5C methylation modification may be an accomplice in colorectal cancer escaping from anti-tumor effects of innate immunity-type I/III interferon. Front Immunol. 2024;15:1512353
210. Ramanjulu JM, Pesiridis GS, Yang J, Concha N, Singhaus R, Zhang SY. et al. Design of amidobenzimidazole STING receptor agonists with systemic activity. Nature. 2018;564:439-43
211. Zhong L, Wu J, Zhou B, Kang J, Wang X, Ye F. et al. ALYREF recruits ELAVL1 to promote colorectal tumorigenesis via facilitating RNA m5C recognition and nuclear export. NPJ Precis Oncol. 2024;8:243
212. Young M, Jackson-Spence F, Beltran L, Day E, Suarez C, Bex A. et al. Renal cell carcinoma. Lancet. 2024;404:476-91
213. Tian J, Gao J, Cheng C, Xu Z, Chen X, Wu Y. et al. NOP2-mediated 5-methylcytosine modification of APOL1 messenger RNA activates PI3K-Akt and facilitates clear cell renal cell carcinoma progression. Int J Biol Sci. 2024;20:4853-71
214. Hopper T, Olabisi OA. APOL1-Mediated Kidney Disease. Jama. 2024;331:1668-9
215. Yang L, Yin H, Chen Y, Pan C, Hang H, Lu Y. et al. Low expression of PEBP1P2 promotes metastasis of clear cell renal cell carcinoma by post-transcriptional regulation of PEBP1 and KLF13 mRNA. Exp Hematol Oncol. 2022;11:87
216. Lamade AM, Wu L, Dar HH, Mentrup HL, Shrivastava IH, Epperly MW. et al. Inactivation of RIP3 kinase sensitizes to 15LOX/PEBP1-mediated ferroptotic death. Redox Biol. 2022;50:102232
217. Manivarma T, Kapralov AA, Samovich SN, Tyurina YY, Tyurin VA, VanDemark AP. et al. Membrane regulation of 15LOX-1/PEBP1 complex prompts the generation of ferroptotic signals, oxygenated PEs. Free Radic Biol Med. 2023;208:458-67
218. Wang N, Chen RX, Deng MH, Wei WS, Zhou ZH, Ning K. et al. m(5)C-dependent cross-regulation between nuclear reader ALYREF and writer NSUN2 promotes urothelial bladder cancer malignancy through facilitating RABL6/TK1 mRNAs splicing and stabilization. Cell Death Dis. 2023;14:139
219. Zhang C, Chang X, Chen D, Yang F, Li Z, Li D. et al. Downregulation of HDGF inhibits the tumorigenesis of bladder cancer cells by inactivating the PI3K-AKT signaling pathway. Cancer Manag Res. 2019;11:7909-23
220. Wang JZ, Zhu W, Han J, Yang X, Zhou R, Lu HC. et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun (Lond). 2021;41:560-75
221. Zhao G, Yuan H, Li Q, Zhang J, Guo Y, Feng T. et al. DDX39B drives colorectal cancer progression by promoting the stability and nuclear translocation of PKM2. Signal Transduct Target Ther. 2022;7:275
222. Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen ZP. et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med. 2013;19:1649-54
223. Zhang Y, Chen XN, Zhang H, Wen JK, Gao HT, Shi B. et al. CDK13 promotes lipid deposition and prostate cancer progression by stimulating NSUN5-mediated m5C modification of ACC1 mRNA. Cell Death Differ. 2023;30:2462-76
224. Cheng X, Xu J, Cui Y, Liu J, Chen Y, He C. et al. Nanovesicles for Lipid Metabolism Reprogram-Enhanced Ferroptosis and Magnetotherapy of Refractory Tumors and Inhibiting Metastasis with Activated Innate Immunity. ACS Nano. 2025;19:7213-30
225. Masetti M, Carriero R, Portale F, Marelli G, Morina N, Pandini M. et al. Lipid-loaded tumor-associated macrophages sustain tumor growth and invasiveness in prostate cancer. J Exp Med. 2022;219:e20210564
226. Zhu W, Wan F, Xu W, Liu Z, Wang J, Zhang H. et al. Positive epigenetic regulation loop between AR and NSUN2 promotes prostate cancer progression. Clin Transl Med. 2022;12:e1028
227. Raychaudhuri R, Lin DW, Montgomery RB. Prostate Cancer: A Review. Jama. 2025;333:1433-46
228. Dai C, Dehm SM, Sharifi N. Targeting the Androgen Signaling Axis in Prostate Cancer. J Clin Oncol. 2023;41:4267-78
229. Ren L, Zhang T, Zhang J. Recent advances in dietary androgen receptor inhibitors. Med Res Rev. 2024;44:1446-500
230. Zhu X, Farsh T, Vis D, Yu I, Li H, Liu T. et al. Genomic and transcriptomic features of androgen receptor signaling inhibitor resistance in metastatic castration-resistant prostate cancer. J Clin Invest. 2024;134:e178604
231. Francoeur AA, Monk BJ, Tewari KS. Treatment advances across the cervical cancer spectrum. Nat Rev Clin Oncol. 2025;22:182-99
232. Yu M, Ni M, Xu F, Liu C, Chen L, Li J. et al. NSUN6-mediated 5-methylcytosine modification of NDRG1 mRNA promotes radioresistance in cervical cancer. Mol Cancer. 2024;23:139
233. Chen Y, Zuo X, Wei Q, Xu J, Liu X, Liu S. et al. Upregulation of LRRC8A by m(5)C modification-mediated mRNA stability suppresses apoptosis and facilitates tumorigenesis in cervical cancer. Int J Biol Sci. 2023;19:691-704
234. Wang L, Zhang J, Su Y, Maimaitiyiming Y, Yang S, Shen Z. et al. Distinct Roles of m(5)C RNA Methyltransferase NSUN2 in Major Gynecologic Cancers. Front Oncol. 2022;12:786266
235. Wang Y, Sun Z, Ping J, Tang J, He B, Chang T. et al. Cell volume controlled by LRRC8A-formed volume-regulated anion channels fine-tunes T cell activation and function. Nat Commun. 2023;14:7075
236. Deneka D, Sawicka M, Lam AKM, Paulino C, Dutzler R. Structure of a volume-regulated anion channel of the LRRC8 family. Nature. 2018;558:254-9
237. Yu D, Chen C, Sun L, Wu S, Tang X, Mei L. et al. KRT13-expressing epithelial cell population predicts better response to chemotherapy and immunotherapy in bladder cancer: Comprehensive evidences based on BCa database. Comput Biol Med. 2023;158:106795
238. Yin L, Li Q, Mrdenovic S, Chu GC, Wu BJ, Bu H. et al. KRT13 promotes stemness and drives metastasis in breast cancer through a plakoglobin/c-Myc signaling pathway. Breast Cancer Res. 2022;24:7
239. Chen SJ, Zhang J, Zhou T, Rao SS, Li Q, Xiao LY. et al. Epigenetically upregulated NSUN2 confers ferroptosis resistance in endometrial cancer via m(5)C modification of SLC7A11 mRNA. Redox Biol. 2024;69:102975
240. Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. 2025;75:10-45
241. Armstrong DK, Alvarez RD, Bakkum-Gamez JN, Barroilhet L, Behbakht K, Berchuck A. et al. Ovarian Cancer, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19:191-226
242. Meng H, Miao H, Zhang Y, Chen T, Yuan L, Wan Y. et al. YBX1 promotes homologous recombination and resistance to platinum-induced stress in ovarian cancer by recognizing m5C modification. Cancer Lett. 2024;597:217064
243. Gao W, Chen L, Lin L, Yang M, Li T, Wei H. et al. SIAH1 reverses chemoresistance in epithelial ovarian cancer via ubiquitination of YBX-1. Oncogenesis. 2022;11:13
244. Smith R, Sellou H, Chapuis C, Huet S, Timinszky G. CHD3 and CHD4 recruitment and chromatin remodeling activity at DNA breaks is promoted by early poly(ADP-ribose)-dependent chromatin relaxation. Nucleic Acids Res. 2018;46:6087-98
245. Wang F, Lin JS, Liu K. Steric control of the reaction of CH stretch-excited CHD3 with chlorine atom. Science. 2011;331:900-3
246. Liu X, Wei Q, Yang C, Zhao H, Xu J, Mobet Y. et al. RNA m(5)C modification upregulates E2F1 expression in a manner dependent on YBX1 phase separation and promotes tumor progression in ovarian cancer. Exp Mol Med. 2024;56:600-15
247. Yang S, Zhou D, Zhang C, Xiang J, Xi X. Function of m(5)C RNA methyltransferase NOP2 in high-grade serous ovarian cancer. Cancer Biol Ther. 2023;24:2263921
248. Caswell-Jin JL, Sun LP, Munoz D, Lu Y, Li Y, Huang H. et al. Analysis of Breast Cancer Mortality in the US-1975 to 2019. Jama. 2024;331:233-41
249. Gradishar WJ, Moran MS, Abraham J, Abramson V, Aft R, Agnese D. et al. Breast Cancer, Version 3.2024, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2024;22:331-57
250. Lei S, Zheng R, Zhang S, Wang S, Chen R, Sun K. et al. Global patterns of breast cancer incidence and mortality: A population-based cancer registry data analysis from 2000 to 2020. Cancer Commun (Lond). 2021;41:1183-94
251. Dai Y, Zhao S, Chen H, Yu W, Fu Z, Cui Y. et al. RNA methylation and breast cancer: insights into m6A, m7G and m5C. Mol Biol Rep. 2024;52:27
252. Zhang X, An K, Ge X, Sun Y, Wei J, Ren W. et al. NSUN2/YBX1 promotes the progression of breast cancer by enhancing HGH1 mRNA stability through m(5)C methylation. Breast Cancer Res. 2024;26:94
253. Tian W, Zhu L, Luo Y, Tang Y, Tan Q, Zou Y. et al. Autophagy Deficiency Induced by SAT1 Potentiates Tumor Progression in Triple-Negative Breast Cancer. Adv Sci (Weinh). 2024;11:e2309903
254. Gu X, Wei S, Lv X. Circulating tumor cells: from new biological insights to clinical practice. Signal Transduct Target Ther. 2024;9:226
255. Tie J, Cohen JD, Lahouel K, Lo SN, Wang Y, Kosmider S. et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med. 2022;386:2261-72
256. Wang G, Li J, Bojmar L, Chen H, Li Z, Tobias GC. et al. Tumour extracellular vesicles and particles induce liver metabolic dysfunction. Nature. 2023;618:374-82
257. Yin H, Huang Z, Niu S, Ming L, Jiang H, Gu L. et al. 5-Methylcytosine (m(5)C) modification in peripheral blood immune cells is a novel non-invasive biomarker for colorectal cancer diagnosis. Front Immunol. 2022;13:967921
258. Lee AC. Epitranscriptomics of cancer microniches. Nat Rev Cancer. 2023;23:189
259. Huang Z, Pan J, Wang H, Du X, Xu Y, Wang Z. et al. Prognostic Significance and Tumor Immune Microenvironment Heterogenicity of m5C RNA Methylation Regulators in Triple-Negative Breast Cancer. Front Cell Dev Biol. 2021;9:657547
260. Modi AD, Zahid H, Southerland AC, Modi DM. Epitranscriptomics and cervical cancer: the emerging role of m(6)A, m(5)C and m(1)A RNA modifications. Expert Rev Mol Med. 2024;26:e20
261. Yu J, Liang LL, Liu J, Liu TT, Li J, Xiu L. et al. Development and Validation of a Novel Gene Signature for Predicting the Prognosis by Identifying m5C Modification Subtypes of Cervical Cancer. Front Genet. 2021;12:733715
262. Yun D, Yang Z, Zhang S, Yang H, Liu D, Grützmann R. et al. An m5C methylation regulator-associated signature predicts prognosis and therapy response in pancreatic cancer. Front Cell Dev Biol. 2022;10:975684
263. Li S, Liu Y, Wu X, Pan M, Zhao H, Hong Y. et al. The m(5)C methyltransferase NSUN2 promotes progression of acute myeloid leukemia by regulating serine metabolism. Cell Rep. 2025;44:115661
264. Wang FH, Zhang XT, Tang L, Wu Q, Cai MY, Li YF. et al. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2023. Cancer Commun (Lond). 2024;44:127-72
265. Huang H, Fang L, Zhu C, Lv J, Xu P, Chen Z. et al. YBX1 promotes 5-Fluorouracil resistance in gastric cancer via m5C-dependent ATG9A mRNA stabilization through autophagy. Oncogene. 2025;44:2357-71
266. Lucas MC, Novoa EM. Long-read sequencing in the era of epigenomics and epitranscriptomics. Nat Methods. 2023;20:25-9
267. Shi CJ, Pang FX, Lei YH, Deng LQ, Pan FZ, Liang ZQ. et al. 5-methylcytosine methylation of MALAT1 promotes resistance to sorafenib in hepatocellular carcinoma through ELAVL1/SLC7A11-mediated ferroptosis. Drug Resist Updat. 2025;78:101181
268. Li Y, Liu H, Li J, Fu C, Jiang B, Chen B. et al. MLLT3 Regulates Melanoma Stemness and Progression by Inhibiting HMGB1 Nuclear Entry and MAGEA1 M(5)C Modification. Adv Sci (Weinh). 2025;12:e2408529
269. Yang X, Tao L, Zhou J, Zhang K, Wang H. Ribosome RNA modification in cancer: Biological functions and therapeutic targets. Sci Bull (Beijing). 2024;69:722-6
270. Chen Z, Zeng C, Yang L, Che Y, Chen M, Sau L. et al. YTHDF2 promotes ATP synthesis and immune evasion in B cell malignancies. Cell. 2025;188:331-51.e30
271. Ma HL, Bizet M, Soares Da Costa C, Murisier F, de Bony EJ, Wang MK. et al. SRSF2 plays an unexpected role as reader of m(5)C on mRNA, linking epitranscriptomics to cancer. Mol Cell. 2023;83:4239-54.e10
272. Barbier AJ, Jiang AY, Zhang P, Wooster R, Anderson DG. The clinical progress of mRNA vaccines and immunotherapies. Nat Biotechnol. 2022;40:840-54
Author contact
![]() Corresponding authors: Yu Zhang, Department of Gynecology, Xiangya Hospital, Central South University, Changsha, China; Gynecological Oncology Research and Engineering Center of Hunan Province, Changsha, China; National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China; Tel.: +86 151 1629 6585; Email: xyzhangyuedu.cn. Lisha Wu, Gynecological Oncology Research and Engineering Center of Hunan Province, Changsha, China; National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China; Institute of Medical Sciences, Xiangya Hospital, Central South University, Changsha, China; Tel.: +86 138 7313 9565; Email: lishawuedu.cn.
Corresponding authors: Yu Zhang, Department of Gynecology, Xiangya Hospital, Central South University, Changsha, China; Gynecological Oncology Research and Engineering Center of Hunan Province, Changsha, China; National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China; Tel.: +86 151 1629 6585; Email: xyzhangyuedu.cn. Lisha Wu, Gynecological Oncology Research and Engineering Center of Hunan Province, Changsha, China; National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China; Institute of Medical Sciences, Xiangya Hospital, Central South University, Changsha, China; Tel.: +86 138 7313 9565; Email: lishawuedu.cn.
 Global reach, higher impact
Global reach, higher impact